Liquid biopsy for monitoring anaplastic lymphoma kinase inhibitors in non-small cell lung cancer: two cases compared
Introduction
Along with breast and colorectal, lung cancer still represents one of the most frequently diagnosed malignancy worldwide, often affected by poor prognosis (1). Despite this, the advent of the targeted drugs has been modifying this serious scenario. However, to fully achieve the ambitious goal of a “personalized” treatment in daily clinical practice, it is mandatory to deveop companion diagnostics, able to trace “druggable” targets in individual cancer patients, throughout the continuum of the care.
In non-small cell lung cancer (NSCLC), 3–7% of patients show echinoderm microtubule associated-protein like 4-anaplastic lymphoma kinase (EML4-ALK) translocation and could be treated with anaplastic lymphoma kinase (ALK)-inhibitors. However, under crizotinib, a first-generation ALK-inhibitor, patients develop drug resistance after a median of 12 months. To overcome crizotinib resistance, several next-generation ALK inhibitors have been developed, including, among others, ceritinib, alectinib, brigatinib and lorlatinib (2). In the USA, the therapeutic decision after detecting new lesions in patients receiving crizotinib varies depending on their location and extent (3). In NSCLC, liquid biopsy allowed important improvements in the detection of the epidermal growth factor receptor (EGFR) mutation (4,5). The ability of liquid biopsy to detect the oncogenic gene/protein fusions is a newly investigated field, and is not routinely applied as of yet.
We here present two cases of NSCLC, both rearranged for EML4-ALK and treated accordingly, which differed by clinical outcome. The two patients showed a different trend of liquid biopsy parameters [namely epithelial cellular adhesion molecule (EpCAM)-positive circulating tumor cells (CTCs), EpCAM low/neg CTCs, EML4-ALK-rearranged CTCs, and cell-free mRNA (cfmRNA)] all of them strongly applying as predictive biomarkers of ALK-inhibitors treatments.
Methods
Patients’ clinical characteristics
Informed consent
The subjects recruited in this study had signed the informed consent forms. The study outcome did not affect the patients’ management, and will not affect the future management of the patient that was censored at the end of this study.
Case 1—presentation
In December 2014, a 70-year-old man, heavy smoker, was hospitalized because of acute pain in the dorsal region. He was diagnosed with a thoracic-abdominal aortic aneurysm. Furthermore, in February 2015, a CT-guided pulmonary biopsy was performed because of multiple pulmonary lesions, and documented a poorly differentiated adenocarcinoma G3, TTF1 pos, p63 and CK6 neg. The molecular characterization reported an EGFR wild type, ALK-rearranged tumor specimen.
With the consent of vascular surgeons, we planned a first-line treatment with carboplatin (AUC 4) and pemetrexed with custom reduction of 75% of the standard dose (375 mg/m2) every 21 days; 3 cycles were tolerated quite well, although we documented anemia (G2) and thrombocytopenia (G1) at the end of the second cycle.
The CT evaluation, on June 2015, documented a progressive disease (PD) of the lung, mediastinal lymph nodes and adrenal metastases, so we proposed a second-line treatment with crizotinib (250 mg BID).
The following CT evaluation (October 2015) showed further PD of the lung, mediastinal lymph nodes and adrenal metastases. Moreover, the excision of a subcutaneous nodule of the right mammary gland demonstrated its metastatic origin from the primary NSCLC. We therefore discontinued crizotinib therapy and ceritinib was required for compassionate use.
After 3 months of ceritinib (750 mg/day), the CT evaluation (February 2016) showed further lung PD, so we proposed a personalized posology and weekly schedule of docetaxel. The patient died in April 2016.
Case 2—presentation
In January 2016, a 50-year-old woman, never smoker, reported tickly cough.
A CT performed in February 2016 showed multiple neoplastic lesions of lung parenchyma; subcarinal and paratracheal lymph nodes were involved.
A CT-guided lung biopsy documented a poorly differentiated G3 adenocarcinoma (TTF1 pos, CK7 pos). The molecular characterization detected an EGFR wild type and ALK-rearranged tumor.
The patient underwent 4 cycles of cisplatin (75 mg/m2) plus pemetrexed (500 mg/m2), every 21 days. The treatment was well tolerated, except for abdominal pain (G2). We documented an initial benefit on coughing.
The CT evaluation (May 2016) documented a stable disease (SD) according to RECIST criteria; hence we planned pemetrexed as maintenance treatment (6 cycles).
At the following evaluation (September 2016), we documented clinical and radiological PD. Based on the molecular characterization of the primary tumor (ALK-rearranged), the patient underwent crizotinib, showing rapid improvement. The treatment has been maintained until now, although the dose was reduced (April 2017) at 250 mg/die, because of neutropenia (G2) and increase of transaminase level (G1).
EpCAM+ CTC detection by CellSearch
We enumerated CTCs in whole blood by the CellSearch System (CS) (Menarini, BO, Italy) according to the manufacturer’s instructions and user’s guidelines (standard assay, SA) (6). Moreover, the FITC-conjugated M30 antibody (clone M30, Vinci Biochem, Vinci, FI, Italy; apoptosis marker) was included in the assay in order to test CTCs viability as previously described (7). Furthermore, in order to investigate the full CK profile of CTCs, we integrated the CTC assay as previously described by using the CXC Kit (Menarini) with FITC-conjugated anti-CK7, -CK14 (clone LP5K and LL002, respectively, Millipore, Billerica, MA, USA), and anti-C11 (clone C11, ACZON, BO, Italy) (expanded assay, EA). The anti-ALK antibody (clone 5A4, AbCam, CB, UK) was included in the expanded assay to detect ALK-positive CTCs (8).
EpCAM low/neg cells isolation and enumeration
EpCAM low/neg cells were collected downstream of the CellSearch processing, using a 5 µm-pore micro-sieve membrane (VyCAP, Deventer, The Netherlands), as previously described (9). The cells on micro-sieve were stained using a cocktail of fluorescently labelled antibodies, namely: FITC-conjugated detecting CK1-10, 13-16, 18, and 19 (AE1/AE3, eBioscience; C11, ACZON; CK7 and -14, Millipore), APC-conjugated anti-CD16 and -CD45, and DAPI for nuclear staining (Invitrogen, Carlsbad, CA, USA). The anti-ALK antibody (AbCam) was also included to detect rearranged cells. Cells were scored as DAPI+CK+CD45−CD16− and ALK+/−, with a minimum cell size of 4 µm × 4 µm, and a nucleus occupying at least 50% of the CK+ cytoplasm.
Circulating cell-free RNA extraction and purification
Plasma was separated from other blood components by a 10 minutes centrifugation at 1,600 g plus a second 10 minutes centrifugation at 16,000 g (to remove any residual blood cells) both at room temperature, and stored at −80°C until cell-free circulating RNA extraction.
The QiaAmp Circulating Nucleic Acid Kit (Qiagen, NW, Germany) was used to extract total circulating nucleic acids from plasma samples following the manufacturer’s instructions. Circulating nucleic acids were eluted in 50–100 µL and then subjected to cell-free circulating RNA purification by the RNeasy Minelute Cleanup kit (Qiagen, NW, Germany), with the protocol recommended in the QiaAmp Circulating Nucleic Acid Kit appendix (DNase treatment plus RNA cleanup). Cell-free circulating RNA was finally diluted in 20 µL of RNase-free water and stored at −80°C.
Reverse transcription and droplet digital PCR
The One-Step RT-ddPCR Advanced Kit for Probes (BioRad, Hercules, CA, USA) was used to combine reverse transcription and droplet digital PCR (RT-ddPCR) for absolute RNA quantification and detection of specific cell-free mRNA transcripts. The expression of 2 of the most common variants of EML4-ALK fusion protein was investigated by a custom synthesized assay (BioRad) slightly modified from what described in Wang et al. [2015] (10) using the following forward primers: EML4-ALK variant 1: AGCCCACACCTGGGAAAGGAC; EML4-ALK variant 3 (detecting both isoform a and b): CTTGGGAAAATTCAGATGATAGCCG, a common reverse primer: AGCTTGCTCAGCTTGTACTCAGGG, and a common probe: FAM-CATGGCTTGCAGCTCCTGGTGCTTCC-IOWA BLACK FQ. The latent transforming growth factor beta binding protein 1 (LTBP1) gene was used as housekeeping and its expression detected by a PrimePCR™ ddPCR™ Gene Expression Probe Assay (Biorad, Unique Assay ID: dHsaCPE5049001, HEX-labeled).
Reactions were performed in a 20 µL reaction mix containing 1× droplet PCR supermix, 20 U/µL Reverse Transcriptase, 15 mM DTT, 250 nM of each probe, 900 nM primers and 5 µL of cell-free circulating RNA. Droplets were generated and analyzed using the QX200 system (BioRad). Amplifications were performed using the following conditions: 1 cycle of 50 °C for 60 min, 1 cycle of 95 °C for 10 min, 40 cycles of 95 °C for 30 seconds and 55 °C for 1 min, 1 cycle of 98 °C for 10 min (ramp rate set to 2 °C per second). Positive-, negative- and no template- controls were included in each run, at least in two replicates. The data were acquired and analysed by QuantaSoft analysis software version 1.7.4 (BioRad).
The specificity of the mutation detection assays was tested by RT-ddPCR of RNA samples harboring the specific translocations, in detail: H2228 RNA for EML4-ALK variant 3 and a certified reference standard for EML4-ALK variant 1 (Horizon, Discovery, CB, UK). Simultaneously, a wild type RNA control was used; this sample never showed false positive mutated droplets.
Results
Both patients (Figures 1,2) received initial platinum-based doublet (CBDCA plus PMTX, and CDDP plus PMTX respectively) chemotherapy (Figures 1A,2A, dotted purple arrow) followed by crizotinib at the time of first documented PD (Figures 1A,2A, continuous purple arrow), because of a documented ALK rearrangement in the primary tumor (Figures 1A,2A, continuous blue arrow). We analyzed EpCAM+ and low/neg CTCs and cfmRNA before the start of chemotherapy (t0) and throughout treatment (subsequent time points), in order to test their potential use as predictors of treatment effectiveness. In the first patient, we repeatedly observed the presence of EpCAM+ CTCs (Figure 1A, green line, EA; t0, t2 and t3) that were partially ALK+ (Figure 1B, t2 and t3). Furthermore, high numbers of EpCAM low/neg CTCs were detected from t0 to t3 (Figure 1A, red line), with ALK+ CTCs ranging from 25% to 44% of this subpopulation (Figure 1C). Conversely, the EpCAM+ CTC SA did not detect any cells at t0 and t1 (Figure 1D).
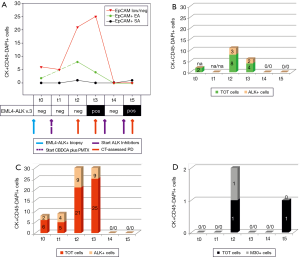
At t2 and t3, after three cycles of CBDCA plus PMTX, when PD was documented by imaging (Figure 1A, red arrow), we found the highest number of EpCAM+ and EpCAM low/neg CTCs, with an ALK+ fraction in both subpopulations (Figure 1B,C). Moreover, at t3 we could reveal for the first time the presence of EML4-ALK cfmRNA (Figure 1A). EpCAM+ SA CTCs were found at t2, when 1 out of 2 cells was apoptotic (M30+) (Figure 1D).
When the patient started crizotinib (t4), EpCAM+ and EpCAM low/neg cells immediately disappeared (Figure 1A, first continuous purple arrow). However, in October the CT revealed a further PD (data not shown), hence the patient switched to ceritinib (Figure 1A, second continuous purple arrow). At t5, we observed a persistence of viable EpCAM+ CTC SA and EML4-ALK cfmRNA (Figure 1A,D, respectively); at this late time point, EpCAM+ EA and EpCAM low/neg were undetectable. The patient rapidly progressed and died in April 2016.
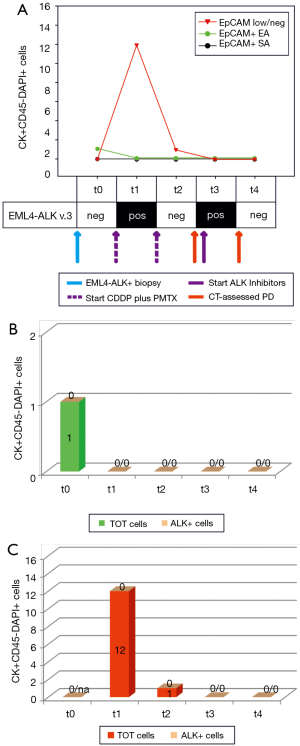
The second case was a never smoker woman, diagnosed with an ALK-rearranged adenocarcinoma (Figure 2A, blue arrow). At baseline (t0), we only observed an EpCAM+ ALK− CTC, with the EpCAM low/neg fraction and cfmRNA being negative. After 4 cycles of CDDP plus PMTX, we noticed that EpCAM+ CTCs had disappeared (Figure 2A,B, EA and SA) while both EpCAM low/neg ALK− CTCs and EML4-ALK cfmRNA were detected (Figure 2A,C, t1). Imaging screening highlighted SD and the patient was treated with pemetrexed only (Figure 2A, second dotted purple arrow). At the subsequent time points, we observed the following: 1 EpCAM low/neg ALK− CTC (t2), EML4-ALK cfmRNA (t3), all markers negative at the last time point (t4). To date, the patient is still showing SD.
Discussion
In ALK-rearranged NSCLC patients, several studies focused on assessing which liquid biopsy marker could be used to detect ALK fusion gene/protein and, in patients treated with ALK TKIs, the onset of resistance.
Consistently with previous reports in other malignancies (11,12), both our case studies demonstrated here that chemotherapy induces the release into circulation of EpCAM low/neg CTCs after only a few cycles. Notably, EpCAM down-modulation is considered the first step in epithelial to mesenchymal transition (EMT), an evolution toward a more aggressive tumor phenotype implicated in resistance to chemotherapy.
In the first case, we found a persistence of ALK+ CTCs in both EpCAM+ and EpCAM low/neg cellular subsets. After starting ALK TKIs, all the CTC subpopulations disappeared, but shortly after, we could detect EML4-ALK cfmRNA and a viable EpCAM+ cell. As suggested by Pailler et al. [2017] (13) the presence of ALK-CTCs after crizotinib is a sign of emerging drug resistance which involves the alteration of pathways other than ALK. Indeed, in our first case EpCAM+ CTCs reemerged after crizotinib and ceritinib, a second generation of ALK TKIs, corresponding to a sudden PD and a worsening of the patient’s conditions. Conversely, in the second patient, we obtained a satisfying stabilization of the disease. In this case, we found CTCs only in the earlier time points. Furthermore, consistently with the imaging documented SD, when the patient was treated with ALK TKIs, we did not observe any CTC subpopulation and EML4-ALK cfmRNA was undetectable.
These two cases are paradigmatic and well depict the heterogeneity in ALK TKIs response observed in clinical practice (14). By combining the biomarkers here proposed, we demonstrated that it is possible to follow the response to treatment and the development of putative drug-resistance mechanisms, to better highlight high-risk patients.
Further in-depth analysis of these biomarkers could open a window onto the cancer behavior and to tailor new therapeutic strategies.
Acknowledgements
We thank Prof. Giuseppe Opocher, Scientific Director of the Veneto Institute of Oncology, IOV-IRCCS, for the new lab facilities provided for the CTC-group at the Institute of Pediatric Research Città della Speranza, IRP, Padova.
Funding: This work was supported by the Italian Ministry of Health (Proposal No: #GR-2010-2303193A “Individualized treatments of patients with advanced NSCLC: potential application for CTCs molecular and phenotypical profiling” to E.R.); and Intramural 5X1000 SINERGIA CTC/cfDNA IOV to R.Z. The work of M Manicone and MC Scaini was supported by IMI [115749] CANCER-ID.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The protocol for this research project has been approved by the Ethics Committee of the institution within which the work was undertaken and it conforms to the provisions in accordance with the Helsinki Declaration as revised in 2013 (CE IOV: 2012/52, 24 September 2012) and written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Awad MM, Shaw AT. ALK inhibitors in non-small cell lung cancer: crizotinib and beyond. Clin Adv Hematol Oncol 2014;12:429-39. [PubMed]
- Bendaly E, Dalal AA, Culver K, et al. Monitoring for and Characterizing Crizotinib Progression: A Chart Review of ALK-Positive Non-Small Cell Lung Cancer Patients. Adv Ther 2017;34:1673-85. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Rossi E, Basso U, Celadin R, et al. M30 neoepitope expression in epithelial cancer: quantification of apoptosis in circulating tumor cells by CellSearch analysis. Clin Cancer Res 2010;16:5233-43. [Crossref] [PubMed]
- Aieta M, Facchinetti A, De Faveri S, et al. Monitoring and Characterization of Circulating Tumor Cells (CTCs) in a Patient With EML4-ALK-Positive Non-Small Cell Lung Cancer (NSCLC). Clin Lung Cancer 2016;17:e173-e177. [Crossref] [PubMed]
- de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015;5:12270. [Crossref] [PubMed]
- Wang Q, Yang X, He Y, et al. Droplet Digital PCR for Absolute Quantification of EML4-ALK Gene Rearrangement in Lung Adenocarcinoma. J Mol Diagn 2015;17:515-20. [Crossref] [PubMed]
- Chebouti I, Kasimir-Bauer S, Buderath P, et al. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget 2017;8:48820-31. [PubMed]
- Pavese JM, Bergan RC. Circulating tumor cells exhibit a biologically aggressive cancer phenotype accompanied by selective resistance to chemotherapy. Cancer Lett 2014;352:179-86. [Crossref] [PubMed]
- Pailler E, Oulhen M, Borget I, et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non-Small Cell Lung Cancer Patients. Cancer Res 2017;77:2222-30. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Crizotinib resistance: implications for therapeutic strategies. Ann Oncol 2016;27 Suppl 3:iii42-iii50. [Crossref] [PubMed]


