MicroRNA expression profile in primary lung cancer cells lines obtained by endobronchial ultrasound transbronchial needle aspiration
Introduction
Lung cancer is the leading cause of death in industrialized nations and is growing in young people and women (1). Despite large studies devised to develop new strategies for early diagnosis to increase chemotherapy response and prognosis, mortality remains high and effective therapies for advanced lung cancer are still lacking (2). A better understanding of lung cancer biology and the discovery of novel cancer biomarkers are paramount to achieve effective therapies especially for patients with advanced non-small cell lung cancer (NSCLC).
Recent studies showed that microRNA (miRNA) could be ideal candidate biomarkers for cancer prognosis since their dysregulation was found to correlate with the onset and progression of several malignancies including lung cancer (3,4). miRNA can be accurately analysed in (formalin-fixed paraffin-embedded) FFPE and in fresh tissue biopsies (5). In addition, miRNA is present in biological fluids (blood, plasma, saliva and urine) and are altered in asymptomatic early stage lung cancer patients (6). Different miRNA expression profiles were found to be associated with different lung cancer subtypes and there is evidence that miRNAs could play a role in targeted therapy in different oncological settings (7-9). Despite the potential clinical value of miRNA signatures, the clinical utility of identified signatures has yet to be demonstrated. Recent studies showed that endobronchial ultrasound transbronchial needle aspiration (EBUS-TBNA) provides adequate cytological specimens for lung cancer diagnosis and staging, including detection of the major genetic alterations identified in lung cancer: epidermal growth factor receptor-tyrosine kinase (EGFR) mutation status, anaplastic lymphoma kinase fusion genes (ALK), and Kirsten ras oncogene homologue (KRAS) mutation status (10).
The present study describes a strategy for high-throughput miRNA expression profile analysis of lung cancer lymph nodal metastasis using primary cell lines established from EBUS-TBNA samples. To validate the EBUS-TBNA miRNA profile, the results were compared with those obtained from mediastinoscopies, the “gold standard” procedure for mediastinal staging in NSCLC patients. FFPE biopsies samples were obtained from mediastinoscopies surgical specimens present in our tissue bank.
Methods
The institutional ethical committee approved this prospective single centre study (registration number: R65/14-IEO76), and informed consent was obtained. Patients with proven lung cancer underwent routine EBUS-TBNA for the diagnosis of suspect lymph node metastasis, and cytological specimens were collected for epithelial cell culture and subsequent transcriptomic miRNA expression analysis.
EBUS-TBNA procedures
EBUS-TBNA was performed under local anaesthesia (1% lidocaine) and moderate sedation provided by an anaesthesiologist with spontaneous ventilation. All procedures were performed by the same team of interventional pulmonologists using a convex-probe (EBUS Convex Probe BF-UC180F; Olympus) and a dedicated ultrasound processor (EU-ME2; Olympus). EBUS-TBNA specimens were collected with a 22 gauge dedicated needle (Vizishot NA-201SX-4022; Olympus).
A very small amount of the aspirated material was pushed out by the internal stylet and smeared onto glass slides for immediate on-site evaluation [rapid on site evaluation (ROSE)]. The remaining aspirate and other needle passages were put into saline solution for cell block processing and histological evaluation. One dedicated needle passage was put into a culture basal medium for primary cell cultures.
Primary cell culture
The dedicated EBUS-TBNA specimen was processed within 30 minutes after the end of the procedure and placed in a sterile falcon filled with 5 mL of cell culture basal medium Ham’s F12/DMEM 1:1 supplemented with 1% foetal bovine serum, 50 ng/mL L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 10 µg/mL gentamicin, 0.5 µg/mL amphotericin B, 10 µg/mL human transferrin, 1 µg/mL human insulin, 1µg/mL hydrocortisone, 10 mM Hepes pH 7.5, 50 µM/L-ascorbic acid, 15 nM sodium selenite, 0.1 mM ethanolamine, and 50 ng/mL cholera toxin. EBUS-TBNA samples were spun down for 5 min at 1,000 g at RT, resuspended in 3 mL of complete medium further supplemented with 10 nM epidermal growth factor EGF, 35 µg/mL bovine pituitary extract, and 10 nM triiodothyronine, and cultured on six-well collagen I-coated plates (Collagen Cellware, Biocoat, Corning, USA) in a humidified incubator with 5% CO2. Primary cells were grown for 6 to 12 days and washed twice with PBS prior to total RNA extraction.
Immunofluorescence
Immunofluorescence was used to check the expression of lung epithelial (CCA, for bronchiolar epithelium, and SP-C, for alveolar epithelium) and neuroendocrine (chromogranin A) markers on EBUS-derived and plated cells. All the following steps were carried out at room temperature. Permeabilization was achieved with 0.2% BSA, 0.1% Triton, and 1× PBS for 10 min, followed by one wash with 1x PBS. Blocking was carried out with 2% BSA for 30 min. Primary antibodies were added and left for 1 h. Following two washes with 1x PBS, secondary antibodies were added for 30 min (light protected), then another two washes with 1× PBS were performed. Post-fixing was achieved with 2% PFA for 1 min, followed by DAPI staining for 5 min and another two washes with 1x PBS. A final post-fixing was done with 2% PFA followed by one final wash with 1× PBS. Slides were mounted and analysed. The following antibodies were used: CCA, goat polyclonal raised against a peptide mapping near the mouse protein C-terminal (CC10 T-18, Santa Cruz Biotechnologies, sc-9772), dilution 1:400; secondary antibody, donkey anti goat Cy3, dilution 1:400; SP-C, rabbit polyclonal raised against a.a. 1–20 from human protein N-terminal (Anti-Prosurfactant Protein C, pro-SP-C, Millipore, AB3786), dilution 1:1,000; secondary antibody, donkey anti rabbit Alexa 647, dilution 1:100; chromogranin A, rabbit polyclonal raised against the human protein C-terminal (Abcam, Ab15160), dilution 1:100; secondary antibody, donkey anti rabbit Alexa 647, dilution 1:100.
RNA/miRNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from EBUS-TBNA primary cells using the AllPrep DNA/RNA/miRNA Universal Kit and from FFPE tissue samples obtained by mediastinoscopy using the AllPrep DNA/RNA FFPE Kit, both protocols automated on QIAcube, according to the manufacturer's instructions (Qiagen, Hilden, Germany).
Following histological assessment of each FFPE tissue block, RNA was extracted from microdissected areas of one to two tissue sections (5–10 µm thick) on glass slides with adequate tumour cellularity (>60%), selected by a pathologist. The total quantity of RNA extracted in each sample (EBUS-TBNA and mediastinoscopies) is shown in Table 1.
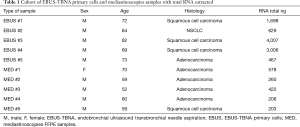
Full table
Total RNA extracted from EBUS-TBNA-derived primary cells was measured using the NanoDrop® ND-1,000 spectrophotometer and reverse transcribed with the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) in 20 µL of final volume and 5 ng of cDNA/reaction were analysed by PCR. Quantitative PCR was performed using UPL probes (Universal ProbeLibrary; Roche, Switzerland) and primers specific for KRT5 and KRT14 (expressed in epithelial basal cells), KRT18 (expressed in bronchial and alveolar epithelium), or PTPRC and PECAM-1 (expressed in endothelial cells), or PDGFRB (expressed in mesenchymal cells) and LCP2 (expressed in lymphocyte), in a final volume of 15 µL of LightCycler 480 Probe Master mix (Roche). UPL probe and primer combinations specific for each target were designed with the free web-based ProbeFinder Software (Roche) and reported in Table S1. Real-time quantitative PCR analysis (RT-qPCR) reaction was run in a LightCycler 480 real-time PCR instrument (Roche) in 96 wells format, using the following thermal cycling conditions: 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s and 72 °C for 60 s and final cooling at 40 °C for 30 s.

Full table
For each mRNA target in each sample, the expression level was measured in triplicate and the average Cq was calculated. Data (average Cq) were normalized to the average Cq value of the two endogenous reference genes (GAPDH and GUSB) using the 2−ΔCq method.
For miRNA expression analysis, 10 ng of total RNA, measured using the Quant-iT™ RiboGreen® RNA assay kit (Thermo Fisher Scientific), were reverse transcribed with Megaplex™ miRNA-specific stem-loop RT Primers Human Pool A v 2.1 (Thermo Fisher Scientific) and TaqMan® MicroRNA reverse transcription kit (Thermo Fisher Scientific) according to the manufacturer’s instructions; 5 µL of reverse transcribed product were pre-amplified for 14 cycles using the TaqMan PreAMP Mastemix and Megaplex PreAMP primers Pool A v 2.1 according to the manufacturer’s instructions (Thermo Fisher Scientific). The PCR reaction was performed using the TaqMan Universal Master Mix II, No AmpErase UNG (Thermo Fisher Scientific) by loading 100 µL of the pre-amplified mixture (final dilution 1:200) in each of the eight lanes of the TaqMan® Low Density Array miRNA Panel A v 2.0 (Thermo Fisher Scientific). Real-Time PCR was carried out on the ViiA7 Real-Time PCR System (Thermo Fisher Scientific) using the manufacturer’s recommended cycling conditions (50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min) and setting an automatic threshold. Cq data of miRNAs were normalized using the RNU6-1 as housekeeping gene. Normalized Cq (Cqn) were calculated as previously described (9). Hierarchical clustering analysis was performed using Cluster 3.0 for Mac OS X (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and Java Treeview (http://jtreeview.sourceforge.net). Spearman rank correlation and centroid clustering methods were used on Cqn data.
Results
The immunofluorescence analysis using known markers of alveolar and neuroendocrine cells constituting the airway epithelium confirmed the lung origin of the established EBUS primary cell lines (Figure 1A).
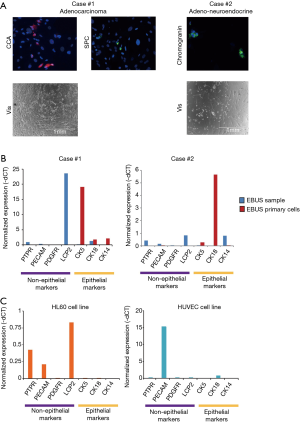
RT-qPCR revealed a high expression of epithelial markers in EBUS-derived primary cell lines which was almost absent in the EBUS-TBNA specimen (Figure 1B), while the EBUS-TBNA samples were positive to the expression of non-epithelial markers (Figure 1B).
As a control, the expression of non-epithelial markers was checked in two commercial cell lines of non-epithelial origin (HL60 and HUVEC) and proved positive (Figure 1C). These data confirmed the successful establishment of lung epithelial cells from EBUS-TBNA specimens.
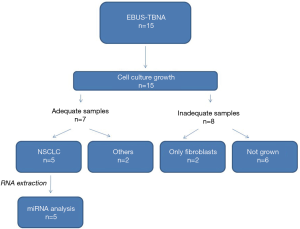
We performed 15 EBUS-TBNA procedures in patients with suspect lymph node NSCLC metastasis for primary cell cultures. Seven out of 15 samples presented an adequate growth on culture and in five patients was possible to extract RNA followed by miRNA profiling analysis. The percentage of adequate growth on culture was about 30% of the total EBUS-TBNA sampled. A flow-chart of the procedure is shown in Figure 2.
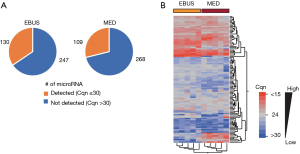
Analysis of the miRNA expression profile of the five independent EBUS-TBNA-derived primary cell lines using TaqMan® Array Human MicroRNA Card A allowed the screening of 377 different human miRNAs; 130 miRNAs (~35% of the total analyzable) were detected (with Cqn ≤30) in all five cell lines. Relative quantities of miRNAs detected in all five EBUS-primary cell lines ranged from 22 to 26 Cqn (i.e., the 25th and 75th quartile intervals) (Figure 3A; Table S2).

Full table
Analysis of the miRNA expression profile of microdissected formalin-fixed lung cancer tissue (“MED” samples; N=5) compared with the profile of EBUS-primary cells (Figure 3A) showed a slight decrease in the average number of miRNA detected (i.e., with Cqn ≤30) in all five FFPE samples (109 vs. 130; Figure 3A). This was probably due to a partial degradation of some miRNA species in FFPE samples.
A total of 102 miRNAs (~78% in EBUS-TBNA primary cell lines and ~94% of FFPE mediastinoscopies) were commonly detected in all samples analysed (with Cqn ≤30), confirming a strong similarity of the two miRNA profiles as also shown in hierarchical cluster analysis (Figure 3B). A list of miRNAs detected in EBUS-primary cells, mediastinoscopy samples and common miRNAs are shown in Table S2.
Discussion
New trends in thoracic oncology are characterized by non-invasive therapies and less-invasive methods for the diagnosis and staging of advanced NSCLC, and targeted therapies (11) designed to reduce patient discomfort and complications and improve survival rates.
Surgical histological specimens have been supplanted by EBUS-TBNA in different clinical scenarios, especially in lung cancer diagnosis and staging (12). In recent years, several studies have demonstrated that EBUS-TBNA reaches the same percentage of diagnosis as surgical biopsies, and allows molecular analyses and mutation detections for target therapies in advanced lung cancer patients (10). Due to its low invasiveness, EBUS-TBNA can also be repeated on “long survival” patients to re-characterize molecular status for personalized treatment.
Different studies have investigated the association between miRNA alterations and lung cancer onset and progression. miRNAs were shown to be prognostic factors, or early diagnostic markers, or more intriguingly, factors determining chemotherapy or biological drug responses (5).
Developing novel biomarkers in conjunction with less invasive methods of diagnosis and staging of NSCLC patients, especially EBUS-TBNA, is paramount to offer patients optimal care with the new perspectives of targeted therapies. The integration of miRNA screening studies with clinical protocols is mandatory to transfer the proposed miRNA biomarkers to the clinical setting. Standardizing methods for collection of biological samples and optimizing miRNA profile analyses, particularly when starting from limited amounts of specimens, represents a definitive strategy to increase the assessment of proposed biomarkers in clinical studies.
As previously reported, miRNAs are resistant to degradation and can be easily identified in FFPE specimens, but few data are available on the feasibility of miRNA expression analyses in cytological specimens. A recent study investigated mRNA and miRNA expression profiles in EBUS-TBNA stored specimens (13). However, it did not address the purity of the samples to derive miRNA/mRNA expression profiles in terms of lung cancer cells fraction, a major issue since miRNAs have been shown to be tissue-specific (14).
The use of primary cell lines instead of the completely fixed cytological specimens collected from EBUS-TBNA has been never described, but should guarantee a higher specificity of the miRNA expression profile, avoiding “contamination” of the neoplastic cell miRNA profile by other non-epithelial cells (lymphocytes, blood and stromal cells) which are abundant in lymph node samples.
This study obtained primary cell cultures from EBUS-TBNA specimens removing most of the non-epithelial cells (lymphocytes, blood and stromal cells) that might affect the gene expression profile of NSCLC cellularity and miRNA results. The results demonstrate the feasibility of obtaining pure populations of primary NSCLC cells from EBUS-TBNA specimens and a complete miRNA expression profile analysis, which strongly correlates with those obtained from microdissected lymph nodal metastases from mediastinoscopy specimens. In addition, the establishment of EBUS-TBNA primary NSCLC cell lines also provides experimental models to test the role of these miRNAs in the biology of lung cancer metastases, which may also have therapeutic indications. Our results are promising for the management of advanced NSCLC patients. The possibility to use EBUS-TBNA specimens for a primary cell culture and whole miRNA expression profiles provides an excellent tool in the personalized therapy of advanced lung cancer patients. On the basis of the present results, we have designed a prospective study to collect EBUS-TBNA primary NSCLC cell lines to evaluate the expression of a miRNA signature profile in stage IIIA NSCLC patients. We plan to evaluate a miRNA signature based on response to chemotherapy that could predict a good prognosis and single out the best candidates for therapy, a crucial point in personalized treatment era.
Acknowledgements
We thank Giuseppina Bonizzi, Francesca Montani, Giovanni Bertalot, Rose Mary Carletti and Stefania Pirroni for technical support. We also thank the Division of Thoracic Surgery, the Department of Pathology, the Biobank for Translational Medicine at IEO and Umberto Veronesi Foundation for the fellowship support to Dr. Roberto Cuttano.
Funding: Fabrizio Bianchi is supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) MFAG17568 and from MIUR (The Italian Ministry of University and Research). Pier Paolo Di Fiore is supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) IGMCO10.000, from MIUR (the Italian Ministry of University and Research) and from the Italian Ministry of Health. Manuela Vecchi is supported by a grant from the Italian Ministry of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The institutional ethical committee approved this prospective single centre study (registration number: R65/14-IEO76), and informed consent was obtained.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the tnm classification for lung cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Nakanishi H, Taccioli C, Palatini J, et al. Loss of miR-125b-1 contributes to head and neck cancer development by dysregulating TACSTD2 and MAPK pathway. Oncogene 2014;33:702-12. [Crossref] [PubMed]
- Li C, Lyu J, Meng QH. MiR-93 Promotes tumorgenesis and metastasis of non-small-cell lung cancer cells by activating the PI3K/Akt pathway via inhibition of LKB1/PTEN/CDKN1A. J Cancer 2017;8:870-9. [Crossref] [PubMed]
- Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430-41. [Crossref] [PubMed]
- Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503. [Crossref] [PubMed]
- Xu H, Cheung IY, Guo HF, et al. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res 2009;69:6275-81. [Crossref] [PubMed]
- Cortez MA, Ivan C, Valdecanas D, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2015;108:djv303. [PubMed]
- Montani F, Marzi MJ, Dezi F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063. [Crossref] [PubMed]
- Casadio C, Guarize J, Donghi S, et al. Molecular testing for targeted therapy in advanced non-small cell lung cancer: suitability of endobronchial ultrasound transbronchial needle aspiration. Am J Clin Pathol 2015;144:629-34. [Crossref] [PubMed]
- Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker testing in lung carcinoma cytology specimens: a perspective from members of the pulmonary pathology society. Arch Pathol Lab Med 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Varela-Lema L, Fernández-Villar A, Ruano-Ravina A. Effectiveness and safety of endobronchial ultrasound-transbronchial needle aspiration: a systematic review. Eur Respir J 2009;33:1156-64. [Crossref] [PubMed]
- Nakajima T, Zamel R, Anayama T, et al. Ribonucleic acid microarray analysis from lymph node samples obtained by endobronchial ultrasonography-guided transbronchial needle aspiration. Ann Thorac Surg 2012;94:2097-101. [Crossref] [PubMed]
- Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016;44:3865-77. [Crossref] [PubMed]


