Identification of proteasome subunit beta type 3 involved in the potential mechanism of corticosteroid protective effectiveness on beta-2 adrenoceptor desensitization by a proteomics approach
Background
Asthma is a chronic inflammatory disease characterized by airway inflammation with mucus hypersecretion and hyperresponsiveness to various nonspecific stimuli (1,2). Corticosteroids are currently the most potent anti-inflammatory agents used to treat chronic inflammatory diseases such as asthma (3). At the same time, they are usually used to prevent β2 adrenoceptor (β2AR) desensitization in clinical and experimental practice (4). As we all known, β2AR agonists are commonly used to relieve symptoms of asthmatic patients, but their desensitization often limits its use in clinical practice. The mechanism of corticosteroids’ protective effect on β2AR desensitization is commonly thought that human β2 receptor gene contains several glucocorticoid response element (GRE) sequences in its promoter region, their stimulation results in an accelerated rate of transcription of the β2AR gene. Theoretically, therefore, use of corticosteroids should up-regulate β2AR expression and make subjects more sensitive to β2AR agonist therapy (5,6). But, besides the above mechanism, there are other reasons or factors involved in the process of corticosteroid protective effect on β2AR desensitization, there is still obscure. And, meanwhile, the relationship of intracellular signaling pathways for β2AR and corticosteroid protective effect on β2AR desensitization is unclear (7). So, a high throughput analysis technology (8)—comparative proteomics is needed to study the difference expression proteins between β2AR desensitization asthmatic mice and corticosteroid-treated β2AR desensitization asthmatic mice. In order to study the potential pathogenesis of corticosteroid protective effect on β2AR desensitization, the β2AR desensitization asthmatic mice model was induced by salbutamol, and DEX was used to establish the β2AR resensitization asthmatic model. After then, their total protein were extracted and separated by two-dimensional gel electrophoresis (2DE), then, the isolated protein spots were compared and analyzed by ImageMaster software and mass spectrometry, and thus proteins were identified. Bioinformatic tools were used to analyze these protein spots and to find related protein spots associated with corticosteroid protective effect on β2AR desensitization. In the end, these protein spots were verified by Western blotting.
Methods
Animals
The study and all procedures concerning animals were approved by the Institutional Animal Care and Use Committee and Conformed to the International Guidelines on the Ethical Use of Animals. Every effort was made to minimize the number of animals used and their suffering. Female BALB/c mice (6-8 weeks old) purchased from Shanghai Laboratory Animal Incorporation, China. Prior to experiment were fed in air-conditioned cages with free access to food and water and maintained on a 12-hour dark/light cycle for a week to acclimatize to their new surroundings.
Antigen sensitization, challenge and treatment
Thirty-two BALB/c (6-8 weeks old) mice were divided into four groups (n=8), which is, group A, control group, PBS-treated; group B, asthmatic group, induced by OVA; group C, β2AR desensitization asthmatic group, treated by OVA and SBT; and group D, corticosteroid-treated β2AR desensitization asthmatic group, treated with OVA, SBT and DEX. Group B, C and D were sensitized on days 0, 14 and 21 by intraperitoneal injection of 200 µg chicken egg OVA (grade V, Sigma, St. Louis, MO, USA) together with 20 mg aluminum hydroxide in a total volume of 200 µL. From the twenty-eighth day on, mice were challenged with an aerosol of 1% OVA (W/V) in saline using an ultrasonic nebulizer 30 min/d for one week. Group C and D underwent the same procedure as group B besides daily intraperitoneal injection of 60 μg salbutamol and inhaling an aerosol of 0.01% salbutamol 30 min/d for a week half an hour before they were challenged with OVA. Meanwhile, group D mice were injected Dexamethasone 5 mg/kg/d for one week by intraperitoneal injection as they were experienced the above same process as Group C. Group A mice were sensitized and challenged with PBS (9).
Measurement of airway hyperreactivity
The mice were anesthetized by intraperitoneal administration of 1% pentobarbital sodium 0.15 mL 24 hours after the last challenge, and then airway hyperresponsiveness to acetylcholine chloride was measured. Each mouse was placed in a plethysmograph chamber and then mechanically ventilated with an animal ventilator (AniRes2005, Beijing SYNOL High-Tech Co. Ltd. China) with a tidal volume of 150 μL and a frequency of 110 breaths per minute. Airway expiration resistance (Re) was continuously measured and recorded for later analysis. After measurement of baseline Re lasting for one minute, the animals were injected with acetylcholine chloride in progressively doses (0.5, 1.5, 5.0, 15, 45 mg/mL) by caudalis vein (35 μL each time) at one minute interval.
Sample preparation
After measurement of airway hyperreactivity, the mice were made bled by retroorbital puncture using heparinized capillary tubes, and blood samples were centrifuged and sera were stored at −70 °C until used. After removal of blood, the mice were used either for collecting bronchoalveolar lavage fluid (BALF) (n=4) or tissue (n=4). The BALF was used for measurement of cytokines. The tracheas of mice were cannulated and lavaged with two 0.8 mL aliquots of PBS. The BALF was centrifuged and supernatants were collected and stored at −70 °C until used. At the same time, the number of mononuclear cells in BALF was counted. After BALF was acquired, the four mice lungs of each group were used to identify the number or β2AR in lung member using a radioligand receptor binding assay (see below). For tissue collection and protein extraction, right lungs of the other four mice of each group were isolated and frozen in liquid nitrogen and stored at −70 °C. When used, one hundred milligram lung tissues of each mouse were taken out and were cut into slices respectively. These slices were transferred to 5 mL steriled centrifuge tubes containing 700 μL of sample buffer [7 mol/L urea, 2 mol/L thiourea, 4% (w/v) Chaps, 40 mmol/L DTT, 2% (v/v) IPG buffer, 1% (v/v) protease inhibitor cocktail]. The mixtures were homogenized using dispersing driver (6,500 r/min, 3-5 times, 3-5 seconds every time, T18 basic ULTRA-TURRAX®, China). Homogenized tissues were incubated for 1 hour at 4 °C, and then sonicated 10 times at an interval setting 15 seconds and working setting 10 seconds. Next, samples were put in ice for 30 minutes and then centrifuged for 30 minutes at 14,000 r/min at 4 °C. Supernatants were transferred to clean vials. Subsequently the protein concentrations were quantified using Bradford assay and stored at −70 °C until used (for 2DE and Western blot analysis). Left lungs were immersed in 10% formaldehyde overnight. Then they were embedded in paraffin, sectioned into 5 mm thickness, and stained with HE. Histologically, the sections were observed under a microscopy by specialists.
Measurement of total IgE, IL-4, and IFN-γ
Total serum IgE, BALF cytokines, interleukin 4 (IL-4) and interferon-gamma (IFN-γ) from each group were measured by standardized sandwich enzyme linked immunosorbent assay (ELISA) according to the manufacturer’s instructions (mlbio, China).
Protein separation and visualization
Proteins (120 μg) from groups C (n=3) and D (n=3) (randomly selected 3 samples from 4 samples) were applied to a 24 cm Immobiline Dry-Strip, pH 3-10 (Amersham Biosciences, Roosendaal, Netherlands), respectively. Strips were rehydrated for 12 hours at 30 V, followed by focusing for 1 hour at 100 V, 1 hour at 500 V, 1 hour at 1,000 V, 1 hour at 5,000 V and subsequently at 8,000 V to 140 kV•h on an IPGPhor (Amersham Biosciences) and stored at −70 °C. Focused IPG strips were equilibrated twice for 15 minutes with gentle shaking in equilibration buffer containing 50 mmol/L Tris-HCl buffer, pH 8.8, 6 mol/L urea, 20% (v/v) glycerol, 0.1% (w/v) sodium dodecyl sulfate (SDS) and 1% (w/v) DTT. In the second equilibration buffer, DTT was replaced with 2.5% (w/v) iodoacetamide in order to remove excess DTT that causes point streaking in silver-stained patterns (10). The equilibrated IPG strips were gently rinsed with distilled water, blotted to remove excess equilibration buffer and then applied onto a 12.5% polyacrylamide gradient gel (26 cm × 20 cm). The Ettan DALT II system (Amersham Biosciences) was used to make the second dimension separation sequentially with a constant voltage of 70 V for one hour, 150 V for two hours, and 400 V for six hours. After SDS-PAGE, the separated gels were visualized by silver staining according to the kit procedures (PlusOne silver staining kit, Amersham Biosciences).
Image analysis
Digitized images of the stained gels were analyzed using the 2-DE gel analysis software ImageMaster 2D ver. 4.0 (Amersham Pharmacia Biotech, Uppsala, Sweden). A comparison report was generated of the qualitative and quantitative differences between the samples for the data according to the changes in intensity (over two times)
In-gel digestion and mass spectrometric analysis
Differentially expressed protein spots were taken from the gels, cut into smaller pieces, and digested with trypsin (Promega, Madison, WI, USA), as described by Gharahdaghi et al. (11) and Joo et al. (12). For matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF-MS) analysis, tryptic peptides were concentrated on POROS R2 columns (Applied Biosystems, Foster City, CA, USA). After successive column washings with 40% methanol, 100% acetonitrile, and 50 mmol/L ammonium bicarbonate, the samples were placed onto the R2 column and eluted in 2 mL of a-cyano-4-hydroxycinnamic acid before MALDI-TOF analysis. The spectra for the protein samples were obtained using a Voyager DE PROMALDI-TOF spectrometer (Applied Biosystems, USA). Protein databases were searched with the MSFit program (http://prospector.ucsf.edu/ucsfhtml3.4/msfit.htm) using monoisotopic peaks. A mass tolerance within 50 per million was allowed for the first analysis; the system was subsequently recalibrated at 20 per million using the list of proteins obtained from the initial analysis. The spectra were also internally calibrated using trypsin autolysis products. The resulting peptide masses were used to search the databases managed by SWISSPROT (http://kr.expasy.org) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Radioligand receptor binding assay
The four mice lung tissues which had been lavaged were respectively minced coarsely with scissors and suspended in 10× volume of 25 mmol/L Tris-HCl buffer (pH 7.4) containing 0.32 mol/L sucrose at 4 °C, and then homogenized with a Polytron homogenizer (Kinematica, Basel, Switzerland) at setting 6 in 30-second bursts. The homogenate was centrifuged at 1,000 ×g for 10 minutes at 4 °C to remove unhomogenized debris, the supernatant was then centrifuged at 40,000 ×g for 20 minutes at 4 °C, and the resulting pellet was washed and recentrifuged at the same speed. The final homogenate was frozen in liquid nitrogen and stored at −80 °C without loss of binding characteristics. Portions of lung membrane at a protein concentration of 10 μg/tube were incubated with (125I) iodocyanopindolol (ICYP) (sp act: 7,400×1,010 Bq/mmol; 3-100 pmol/L) in the presence or absence of excess Iso (200 μmol/L) in 25 mmol/L Tris-HCl buffer (pH 7.4) containing 154 mmol/L NaCl and 1.1 mmol/L ascorbic acid (to prevent oxidation of Iso) in a final volume of 250 μL. The density of β1-receptors was analyzed by ICYP saturation binding in the presence of 0.1 μmol/L ICI 118551, a β2-selective antagonist, a concentration at which practically all β2-receptors were occupied. The density of β2-receptors was analyzed by ICYP saturation binding in the presence of 0.1 μmol/L CGP 20712 A, a β1-selective antagonist, a concentration at which practically all β1-receptors were occupied. Incubation was carried out at 37 °C for 120 minutes, which was found to be optimal for specific binding. Incubations were performed in triplicate. The incubation was terminated by rapid filtration through GF/C glass-fiber filters (Whatman Inc., Clifton, NJ, USA). Each filter was rapidly washed with 3×5 mL ice-cold 25 mmol/L Tris-HCl buffer (pH 7.4). The filters were counted in the Auto Gamma Counting System at an efficiency of 80%. Specific binding was calculated by subtracting nonspecific binding from total binding. Protein concentration was determined by the method of Lowry with bovine serum albumin as a standard.
Immunoblotting
Samples of denatured protein (40 μg) (n=3, the same 3 samples as those of 2-DE, samples from groups A, B, C and D) were separated on 10% SDS-polyacrylamide gel and transferred to nitrocellulose membranes. Blots were blocked in 5% milk in 1× TBST (Tris-buffered saline, 0.5% Tween 20) at 4 °C overnight, and probed with anti-β2AR antibody (1:500 dilution, SC-9042 Santa Cruz Biotechnology, USA) and appropriate secondary antibody conjugated to horseradish peroxidase (1:2,000 dilution, Abcam, UK) at room temperature for two hours, respectively. Meanwhile, the membranes were stripped and reprobed with anti-α-tubulin antibody to confirm equal amounts of loaded samples. Immunoblots of protein bands were visualized with an enhanced chemiluminescence detection kit (Amersham Life Sciences, USA). At the same time, in order to confirm the down-regulation of proteasome subunit beta type 3 identified by 2-DE, the expression of these lung tissue proteins was also examined using Western blotting referred to the above method.
Statistical analysis
Values are expressed as the mean ± standard deviation (SD). Differences among the control group (A), asthmatic group (B), β2AR desensitization asthmatic group (C) and corticosteroid-treated β2AR desensitization asthmatic group (D) were determined by one-way ANOVA. The different protein expression levels of β2AR desensitization asthmatic group (C) and corticosteroid-treated β2AR desensitization asthmatic group (D) were analyzed using Student’s t test. The critical level for significance was set at P<0.05.
Results
Construction of asthmatic mouse model and β2AR desensitization asthmatic mouse model
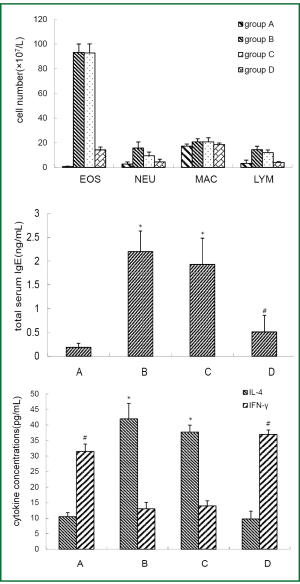
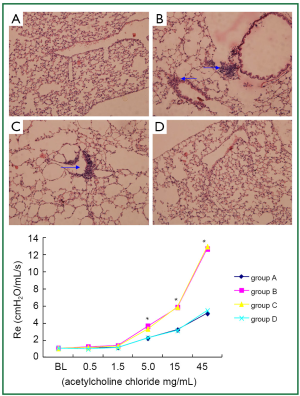
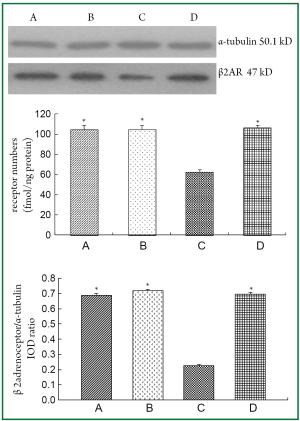
BAL fluid and blood were collected 24 hours after the last OVA aerosol challenge. Eosinophil, neutrophil and lymphocyte counts in the BAL fluid and total serum IgE level were increased (P<0.01) in asthmatic group and β2AR desensitization asthmatic group compared to control group and corticosteroid-treated β2AR desensitization asthmatic group (Figure 1). IL-4 concentrations in BALF of asthmatic group and β2AR desensitization asthmatic group were higher (P<0.01) than those of control group and corticosteroid-treated β2AR desensitization asthmatic group, but IFN-γ concentrations were lower (P<0.01) than those of control group and corticosteroid-treated β2AR desensitization asthmatic group (Figure 1). Airway resistance, determined by an animal ventilator was markedly increased (P<0.01) in asthmatic group and β2AR desensitization asthmatic group compared to control group and corticosteroid-treated β2AR desensitization asthmatic group (Figure 2). In addition, marked infiltration of inflammatory cells, especially eosinophils, into the lamina propria, perivascular, and peribronchiolar areas was found in asthmatic group and β2AR desensitization asthmatic group, compared to control group and corticosteroid-treated β2AR desensitization asthmatic group (Figure 2). Furthermore, β2AR number of β2AR desensitization asthmatic group was obviously lower than that of other three groups (Figure 3). Western blot results showed β2AR total amount changes in control group, asthmatic group, β2AR desensitization asthmatic group, and corticosteroid-treated β2AR desensitization asthmatic group, and α-tubulin was used as internal control (Figure 3). With inflammatory cell count, cytokine concentration of BALF, pathological sections, total serum IgE, airway resistance, membrane receptor number and β2AR total amount changes, asthmatic mouse model and β2AR desensitization asthmatic mouse model were successfully established.
Effect of DEX on β2AR desensitization
Protective effect of DEX on β2AR desensitization was evaluated by radioligand receptor binding assay and Western blot analysis (Figure 3). These results demonstrated that DEX had played an important protective role on β2AR desensitization, so β2AR desensitization asthmatic group and corticosteroid-treated β2AR desensitization asthmatic group were selected for 2-DE in order to find the key protein spots associated with DEX protective role on β2AR desensitization.
2-DE analysis and identification of global differentially expressed protein in group C and group D
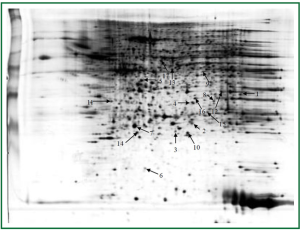
Lungs were isolated from each mouse 24 hours after the last challenge. Proteins from group C and group D (three samples per group) were resolved on 2-D gels and visualized by silver staining (Figure 4). The six gels were analyzed using the 2-DE gel analysis software ImageMaster 2D ver. 4.0 (Figure 5). Then MALDI-TOF MS was used to identify all the differentially expressed protein spots between the two groups and the results were enumerated in Table 1.

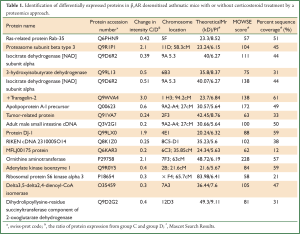
Full table
Key spot verified by western blotting
To verify the 2-DE findings, immunoblotting analysis was made on the key protein spot, that is, proteasome subunit beta type 3. Western blot analysis revealed a significant decrease in proteasome subunit beta type 3 protein level in group D compared to group C (Figure 6), and α-tubulin was used as internal control.

Discussion
We present the comparative proteome analysis of mouse lung tissue from group C and group D. The pH ranges used for the first-dimension gel electrophoresis in this study (3-10) separated a proportion of soluble proteins in the lung tissues (13). To avoid individual and experimental variation, we produced a large number of replicate gels from the same sample or treatment group in order to obtain statistically significant changes in protein levels between the different groups, i.e., β2AR desensitization asthmatic group versus corticosteroid-treated β2AR desensitization asthmatic group (14). Using 2-DE, we found 17 proteins that were differentially expressed in the lung tissues of corticosteroid-treated β2AR desensitization asthmatic group. Four protein spots were down-regulated and 13 protein spots were up-regulated compared to β2AR desensitization asthmatic group. These protein spots were identified by in-gel digestion and MALDI-TOF MS.
In our study four groups were designed: group A, control group, PBS-treated; group B, asthmatic group, induced by OVA; group C, β2AR desensitization asthmatic group, treated by OVA and SBT and group D, corticosteroid-treated β2AR desensitization asthmatic group, treated with OVA, SBT and DEX. We presumed that number of β2AR was not altered in group A, group B and group D but decreased significantly in group C. This finding was proved by the results of our experiments. Meanwhile, group C and group D were selected for 2-DE analysis, attempting to find key protein spots related to corticosteroid protection effect on β2AR desensitization.
Why was β2AR desensitization asthmatic animal model established but not β2AR desensitization animal model established? Because β2AR desensitization asthmatic patients are much more common in clinical practice, and β2AR agonists were usually used to deal with asthmatic patients but not to treat healthy adults (15). Study on β2AR desensitization asthmatic animal model is more significant than just study on β2AR desensitization animal model. Thus, pathogenesis study of corticosteroid protective effect on β2AR desensitization asthmatic mice was designed.
Salbutamol is reportedly an effective bronchodilator used to treat asthmatic patients, but it can also induce β2AR desensitization as other β2AR agonists (16). In this study we found that salbutamol could lead to β2AR desensitization in a murine model of acute asthma by measuring number of membrane β2AR (radioligand receptor binding assay) and total amount of β2AR receptor (Western blot analysis). And we also found that DEX had played a vital protective effect on β2AR desensitization. To investigate the mechanism, we used a proteomic approach to identify the differential expressed proteins between the asthmatic mice with and without DEX intervention
Of those proteins responsible for the protective effects of DEX on β2AR desensitization
Seventeen protein spots were found different expression between group C and group D, of which, 4 protein spots were down-regulated and 13 protein spots were up-regulated compared to group C. Interestingly, we found that Proteasome subunit beta type 3 was up-regulated in β2AR desensitization group, while in other groups, no significant level change was observed, and this was further confirmed by Western blot.
As we all known, a short-term exposure (up to one hour) leads to internalization or redistribution of the phosphorylated receptors away from the cell membrane into endocytic compartments in which they are dephosphorylated and then recycled to the cell membrane. More prolonged exposure (hours to days) leads to receptor degradation or down-regulation, an actual decline in the total cellular receptor number. During the process of β2AR recycling and degradation, proteasome is involved and play a vital role, that is, proteasome might be the reason for β2AR desensitization (17,18). In our study, we found that proteasome subunit beta type 3 was up-regulated only in β2AR desensitized asthmatic group, and when DEX was administrated, it came back to its normal level. Therefore, our results may suggest that the dynamic change of proteasome subunit beta type 3 may be responsible for the process of β2AR desensitization and resensitization.
Proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH. The proteasome has an ATP-dependent proteolytic activity (19), And in our experiment, the increased expression of proteasome subunit may indicate that the proteasome might be involved in the process of salbutamol induced β2AR desensitization. In addition, DEX, not only by increasing the expression of β2AR in gene and protein level, but also decreasing the proteasome system activity to protect the β2AR from proteolytic.
From the above discussion, we can find our study is just a superficial and shallow work, but to our gratified, although a large amount of experiments must be done to further verify our viewpoint, a new target protein related to corticosteroid protective role on β2AR desensitization may have been found after all.
All protein spots are listed in Table 1. The functions of the remaining protein spots will not be deeply discussed because they may not play a major role in our experiment and in our hypothesis according to our bioinformatics search (20-23).
Conclusions
During the process of corticosteroid protective effect on β2AR desensitization, corticosteroids play the vital role not only by traditional way, that is, accelerating β2AR gene transcription rate but also by the decline of proteasome, accurately, proteasome subunit beta type 3.
In summary, we used 2-DE to identify proteins that are differentially expressed in β2AR desensitization asthmatic mice group with and without DEX treatment.
Our data suggest that the identified proteins may provide tools for gauging the success of therapeutic interventions designed to treat β2AR desensitized asthmatic patients. On the other hand, although we identified the altered protein spots in response to corticosteroid protective role on β2AR desensitization, Additional studies are needed to characterize the precise mechanism of action.
Acknowledgements
This study was supported by Natural Science Foundation of China (NO: 30971306), Six big talent peak in Jiangsu province project (the seventh batch NO: 033), Nantong social development project (NO: S2009023) and Nantong fourth period “226 high-level personnel training project” project.
Disclosure: The authors declare no conflict of interest.
References
- Slater M, Rivett DW, Williams L, et al. The impact of azithromycin therapy on the airway microbiota in asthma. Thorax 2013. [Epub ahead of print]. [PubMed]
- Gordon BR. Should vitamin d supplementation be a regular part of asthma care? Otolaryngol Clin North Am 2014;47:97-108. [PubMed]
- Adimi Naghan P, Fahimi F, Nadji SA, et al. A pilot study of polymorphism of adrenergic Beta-2 receptor and mild asthma: a clinical and pharmacogenetic study. Iran J Pharm Res 2013;12:199-204. [PubMed]
- Liu H, Yin KS. Progress of beta-2-adrenergic receptor desensitization and effect of corticosteroids on it. Int J Respir (Chinese) 2007;27:830-3.
- Billington CK, Ojo OO, Penn RB, et al. cAMP regulation of airway smooth muscle function. Pulm Pharmacol Ther 2013;26:112-20. [PubMed]
- Cooper PR, Kurten RC, Zhang J, et al. Formoterol and salmeterol induce a similar degree of β2-adrenoceptor tolerance in human small airways but via different mechanisms. Br J Pharmacol 2011;163:521-32. [PubMed]
- Adner M, Larsson B, Säfholm J, et al. Budesonide prevents cytokine-induced decrease of the relaxant responses to formoterol and terbutaline, but not to salmeterol, in mouse trachea. J Pharmacol Exp Ther 2010;333:273-80. [PubMed]
- Smiley R, Bailey J, Sethuraman M, et al. Comparative proteomics analysis of sarcosine insoluble outer membrane proteins from clarithromycin resistant and sensitive strains of Helicobacter pylori. J Microbiol 2013;51:612-8. [PubMed]
- Liu H, Zhou LF, Zhang Q, et al. Increased RhoGDI2 and peroxiredoxin 5 levels in asthmatic murine model of beta2-adrenoceptor desensitization: a proteomics approach. Chin Med J (Engl) 2008;121:355-62. [PubMed]
- Jang HS, Oh CK, Jo JH, et al. Detection of telomerase activity in psoriasis lesional skin and correlation with Ki-67 expression and suppression by retinoic acid. J Korean Med Sci 2001;16:623-9. [PubMed]
- Gharahdaghi F, Weinberg CR, Meagher DA, et al. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 1999;20:601-5. [PubMed]
- Joo WA, Kang MJ, Son WK, et al. Monitoring protein expression by proteomics: human plasma exposed to benzene. Proteomics 2003;3:2402-11. [PubMed]
- Zhou X, Wang K, Lv D, et al. Global Analysis of Differentially Expressed Genes and Proteins in the Wheat Callus Infected by Agrobacterium tumefaciens. PLoS One 2013;8:e79390. [PubMed]
- Rabilloud T, Lescuyer P. The proteomic to biology inference, a frequently overlooked concern in the interpretation of proteomic data: a plea for functional validation. Proteomics 2013. [Epub ahead of print]. [PubMed]
- Patel M, Pilcher J, Travers J, et al. Use of metered-dose inhaler electronic monitoring in a real-world asthma randomized controlled trial. J Allergy Clin Immunol Pract 2013;1:83-91. [PubMed]
- Cazzola M, Page CP, Rogliani P, et al. β2-agonist therapy in lung disease. Am J Respir Crit Care Med 2013;187:690-6. [PubMed]
- Shenoy SK, McDonald PH, Kohout TA, et al. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science 2001;294:1307-13. [PubMed]
- Hanania NA, Moore RH. Anti-inflammatory activities of beta2-agonists. Curr Drug Targets Inflamm Allergy 2004;3:271-7. [PubMed]
- Benanti JA. Coordination of cell growth and division by the ubiquitin-proteasome system. Semin Cell Dev Biol 2012;23:492-8. [PubMed]
- Sato M, Sato K, Liou W, et al. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J 2008;27:1183-96. [PubMed]
- Kang S, Kim EY, Bahn YJ, et al. A proteomic analysis of the effect of MAPK pathway activation on L-glutamate-induced neuronal cell death. Cell Mol Biol Lett 2007;12:139-47. [PubMed]
- da Costa CA. DJ-1: a newcomer in Parkinson’s disease pathology. Curr Mol Med 2007;7:650-7. [PubMed]
- Thomas B, Beal MF. Parkinson’s disease. Hum Mol Genet 2007;16 Spec No. 2:R183-94.


