A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram
Introduction
Lung cancer is the leading cause of cancer deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for about 85% of all lung cancers (1). Radical resection, including complete tumor resection and systematic lymph node (LN) dissection, is the standard treatment for I–IIIB stage NSCLC. LN metastasis (LNM) is an important factor affecting prognosis: 5-year survival with LNM is 26–53% (2). Therefore, noninvasive preoperative evaluation of LNM is important for lung cancer staging, surgical planning, and prognosis.
In current practice, CT evaluation of LNM relies on morphological changes, including size, shape, presence of necrosis, and external invasion of capsule. The accuracy of these indicators is low (3). Integrated PET/CT is a relatively accurate noninvasive imaging technique, with relatively high specificity for LN staging in patients with lung cancer. However, its higher examination fees and low prevalence limit their clinical application. Recently, radiomics has attracted increased attention of radiologists and clinicians because of its quantitative advantages. Radiomics uses a large number of automated data characterization algorithms to transform image data from a region of interest into the quantitative high-throughput feature space (4). Radiomics features can reflect biological information regarding the tumor, such as cell morphology, molecular and gene expression, which can noninvasively provide information regarding diagnosis, evaluation of prognosis, and prediction of treatment response (5).
Radiomics analysis of primary lesions in colorectal cancer, bladder cancer, and breast cancer predicts the potential for LNM, and has higher sensitivity and specificity than do conventional evaluation methods (6-8). Adenocarcinoma (ADC) is the most common histological subtype of lung cancer. As compared to sub-solid ADC, patients with solid ADC are more likely to have LNM and poor prognosis. To our knowledge, there have been no studies regarding prediction of LNM in primary lung cancer using radiomics features. We hypothesize that a radiomics approach will achieve high accuracy for prediction of LNM in solid lung ADC. As opposed to previous radiomics approaches that require manual contour of lesions, we apply deep learning-driven automatic lesion segmentation for radiomics feature extraction. The purpose of this study was to develop and validate a deep learning masked radiomics nomogram model for the prediction of LNM in solid lung ADC.
Methods
Patients
This retrospective study was approved by our institutional review board. Medical record review was performed in accordance with institutional ethics review board guidelines.
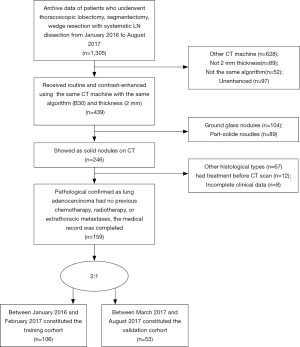
Inclusion criteria were as follows: (I) there was pathological diagnosis of ADC conforming to the 2015 WHO classification of lung cancer; (II) patients underwent thoracoscopic lobectomy, segmentectomy, wedge resection with systematic LN dissection; (III) the medical record contained definitive data regarding pathological type and metastasis to dissected LNs (intrapulmonary, hilar and mediastinal); (IV) within two weeks of surgery, all patients underwent routine and contrast-enhanced CT of the entire thorax using the same CT machine with the same algorithm (B30) and thickness (2 mm); (V) solid nodule appears as a rounded or irregular soft-tissue attenuation on CT; (VI) there was no previous chemotherapy, radiotherapy, or extrathoracic metastases; (VII) laboratory analysis of routine tumor markers were detected within a week before surgery, including CEA, CA125 and CA153. The positive threshold values for CEA, CA125, CA153, were >5, >35 and >25 ng/mL according to the normal ranges used at our institution, respectively.
Exclusion criteria were as follows: (I) clinical data were incomplete, or statistical analysis could not be performed; (II) there was a history of other malignancies or combined malignancies; (III) CT imaging was reconstructed using different algorithms, thicknesses, or reconstruction was performed on a different CT machine.
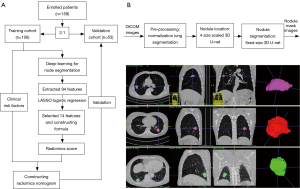
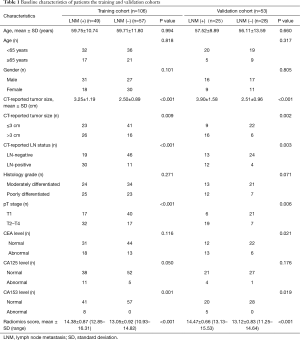
According to the inclusion and exclusion criteria (Supplementary file 1, Figure S1), we limited patient participation between January 2016 and August 2017 at our hospital. We divided the patients into two independent cohorts: 106 patients treated between January 2016 and February 2017 constituted the training cohort, and 53 patients treated between March 2017 and August 2017 constituted the validation cohort. A total of 159 patients were enrolled, including 91 males and 68 females, age 22–81 years (mean 58.76±11.41 years).
Other clinical data, such as surgical records, pathological diagnoses, and LN dissection results, were retrieved from medical records. For review of LNs, CT images were observed on a PACS workstation. The window width and level were adjusted to optimize the examination of mediastinal and hilar LNs. We used multiple criteria for the evaluation of LN status on CT. Negative (benign) criteria were as follows: nodular calcification and intranodal fat tissue; positive (malignant) criteria were as follows: short-axis diameter more than 10 mm, focal low density suggesting necrosis, surrounding fat infiltration suggesting extrafascial extension, and convex margin in hilar LNs (9). The longest dimension of the primary tumor was measured as CT-reported tumor size. A radiologist with 20 years of experience in thoracic CT imaging (Y Guan) and a radiologist with 10 years of experience (X Yang) independently assessed CT-reported LN status and tumor size. Final decisions were reached by consensus. Preoperative clinical stages were determined according to the most recent international staging criteria (8th edition) for lung cancer.
CT image acquisition
All patients received routine and contrast-enhanced CT of the entire thorax, in a multi-detector CT system (Definition AS+ 128-Slice; Siemens Healthcare, Germany). CT scan parameters were as follows: tube voltage, 120 kV; automatic tube current modulation (35–90 mAs); pitch, 0.9; field of view, 180 mm × 180 mm; matrix, 512×512; reconstructed slice thickness 2 mm, and slice increment 2 mm. After routine non-enhanced CT, venous-phase scans started 35 seconds after the contrast media reached 100 HU. Contrast medium (300 mg/mL, iopamidol injection, Bracco) was administered with a dose of 2 mL/kg body weight, at a rate of 3.0 or 3.5 mL/s. All images are exported in DICOM format for image feature extraction.
Lesion segmentation and radiomics features extraction
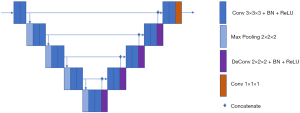
For nodule segmentation, we employed a 3D U-net model (10,11), trained with lung image database consortium (LIDC) datasets (Supplementary file 2, Figure S2). A total of 94 radiomics features (Supplementary file 3, Table S1) were extracted from venous-phase CT images using Pyradiomics, an open source radiomics extraction toolkit (12). The composition of radiomics features and the reasons for selection of venous-phase CT images are described in the supplementary data.

Full table
LN status–related feature selection and radiomics score mode construction
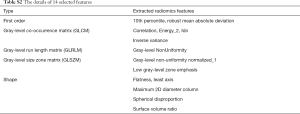
We used the least absolute shrinkage and selection operator (LASSO) logistic regression algorithm to select significant LN status features, with non-zero coefficients from among the 94 imaging features (Supplementary file 3, Table S1), within the training cohort. A formula was generated using a linear combination of selected features that were weighted by their respective LASSO coefficients. A radiomics score was calculated for each patient by the formula to reflect the risk of LNM. The predictive accuracy of the radiomics signature was quantified by the area under curve (AUC) of receiver operator characteristic (ROC) curve in both the training and validation cohorts.
Development of individualized radiomics nomogram
Multivariate logistic regression was used to evaluate the significant risk factors for prediction of LNM from radiomics score, age, CT-reported tumor size, CT-reported LN status, CEA, CA125 and CA153. The variance inflation factor (VIF) was used for collinearity diagnosis in the multivariate logistic regression analysis. Backward step-wise selection was then applied using the likelihood ratio test with Akaike’s information criterion as the stop rule. Based on the multivariate logistic regression analysis, a radiomics nomogram was constructed to provide a quantitative tool that could predict the probability of individual LNM.
The calibration curve was constructed to assess the nomogram. The Hosmer-Lemeshow test was performed to evaluate the goodness-of-fit of the nomogram, and the AUC was calculated to quantify the discriminating performance of the nomogram.
Validation of the individualized radiomics nomogram
The validation cohort was used for internal validation of the radiomics nomogram. The logistic regression formula formed in the training cohort was applied to all patients in the validation cohort. The radiomics score and total points were calculated for each patient. Finally, the calibration was performed and the AUC was calculated.
Clinical application value of the individualized radiomics nomogram
To evaluate the clinical application value of the radiomics nomogram, decision curve analysis (DCA) was conducted by quantifying the net benefits for a range of threshold probabilities in both the training and the validation cohorts.
Statistical analysis
We used the LASSO logistic regression model with penalty parameter tuning that had been conducted using 10-fold cross-validation based on minimum criteria. The likelihood ratio test with backward step-down selection was applied to the multivariate logistic regression model. Detailed descriptions of the LASSO algorithm and DCA are provided in the supplementary data (Supplementary files 4,5).
R statistical software (v3.4, Bell Laboratories, Murray Hill, NJ, USA) was used for all statistical tests. We used the “glmnet” package to perform the LASSO logistic regression model analysis. The VIFs were calculated using the “car” package. The ROC curves were plotted using the “pROC” package. Nomogram construction and calibration plots were performed using the “rms” package. The Hosmer-Lemeshow test was performed using the “generalhoslem” package, to quantify the discrimination performance of the radiomics nomogram. DCA was performed using “dca.R.”. A two-sided P<0.05 was considered statistically significant.
Results
Characteristics of patients in the training and the validation cohorts
The study flowchart and automatic segmentation-based deep learning technique is shown in Figure 1. The characteristics of patients in both the training and the validation cohorts is displayed in Table 1. There was no difference in LNM rate between the two cohorts, with 46.2% (49/106) in the training cohort and 47.2% (25/53) in the validation cohort, respectively (P=0.91). In the combined cohort, 41.9% (31/74) of the pathologically LN (pN) positive patients were understaged, and 17.6% (15/85) of the pathologically-negative LN patients were overstaged as clinical LN (cN) positive according to CT evaluation.
Full table
Selection of features and construction of the radiomics signature
A total of 94 features were extracted from the venous-phase CT images of the training cohort. We screened these features and chose the 14 that had non-zero coefficients (Supplementary file 6, Table S2) as potential predictors using the LASSO logistic regression model (Figure 2A,B). The calculation formula for the radiomics score (Supplementary file 7) based on the selected features is presented in the supplementary data. The radiomics score for LN-positive patients was higher than that of negative LNM patients in the training cohort [(14.38±0.87) vs. (13.05±0.92), P<0.001]. This was then confirmed in the validation cohort [(14.47±0.66) vs. (13.12±0.83), P<0.001]. The radiomics signature also showed a favorable predictive efficacy, with an AUC of 0.854 in the training cohort [95% confidence interval (CI), 0.784–0.924, sensitivity =77.55%, specificity =78.94%.] and 0.803 in the validation cohort (95% CI, 0.683–0.923, sensitivity =72.00%, specificity =85.71%) (Figure 2C,D).
Full table
Development of an individualized nomogram
The univariate logistic regression analysis identified independent predictors, including age, CT-reported tumor size, CT-reported LN status, CEA, CA125, CA153 and radiomics score (Table 2). The radiomics score and CT-reported LN status were recognized as independent predictors for LNM in primary lung ADC patients, using the multivariate logistic regression model (Table 2). The VIFs of the seven predictors mentioned above varied from 1.043 to 1.833, suggesting no collinearity in the diagnosis.

Full table
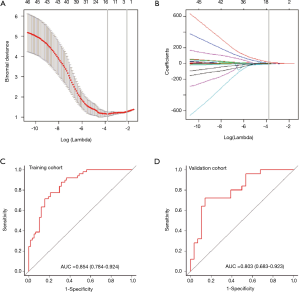
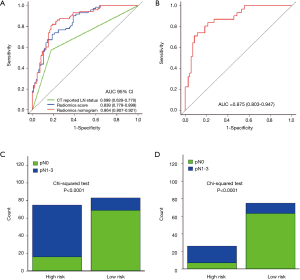
We then constructed a model including radiomics score and CT-reported LN status and used it as nomogram to predict the probability of LNM in the training cohort (Figure 3A). The calibration curve of the radiomics nomogram demonstrated good agreement with the training cohort (Figure 3B). There was no statistical significance in the Hosmer-Lemeshow test (P=0.368). We then confirmed the favorable calibration of the radiomics nomogram in the validation cohort (Figure 3C). The Hosmer-Lemeshow test showed no statistical significance (P=0.138). The AUC of radiomics nomogram in the training cohort was 0.871 (95% CI, 0.804–0.937; sensitivity =85.71%, specificity =77.19%). In the validation cohort, AUC was 0.856 (95% CI, 0.745–0.966; sensitivity =91.66%, specificity =82.14%) (Figure 3D,E). Hence, the radiomics nomogram showed good performance in the training and validation cohorts.
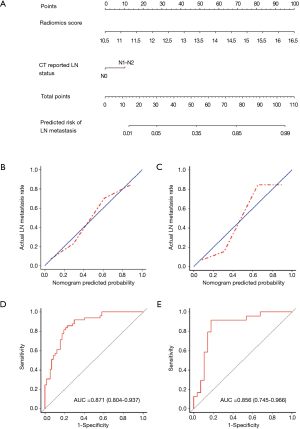
DCA is shown in Figure 4. We found that if the threshold probability of a patient was between 0.46 and 0.83, the radiomics nomogram for predicting LNM would be more beneficial than the strategies “treat all” or “treat none”.
In addition, we evaluated the discriminatory efficiency of the radiomics nomogram in 159 patients and in the LN-negative (cN0) subgroup (n=101) using ROC analysis. The comparison of the discriminatory accuracy between the radiomics nomogram and the radiomics signatures and CT-reported LN status alone is shown in Figure 5A. We found that the AUC of the radiomics nomogram was 0.864 (95% CI, 0.807–0.921), suggesting that the nomogram provided better predictive efficacy. The discriminatory ability in the cN0 subgroup was also confirmed to be good (AUC, 0.875; 95% CI, 0.803–0.947, Figure 5B). After obtaining the risk score, we then defined an optimal risk score cutoff value of 0.42 on the basis of the maximum Youden index in the training cohort, and divided the patients into low-risk and high-risk groups. Importantly, patients in the high-risk group showed a greater possibility for LNM in all patients and in the cN0 subgroup (Figure 5C,D). Meanwhile, nine patients with CT-reported LN positivity were found to have reactive hyperplasia by histopathology. This was consistent with the prediction outcome of the radiomics nomogram.
Discussion
The International Association for the Study of Lung Cancer (IASLC), based on a newly established large database, showed that cN and pN status were closely related to 5-year survival rates (13). Systematic dissection of LNs in lung cancer patients has been widely accepted, but the extent of LN dissection has been the focus of controversy (14-16). Studies (14) have shown that extent of LN resection does not increase peri-operative morbidity or mortality after surgery for stage I lung cancer in the elderly. Some investigators (16) recommend a cutoff of 16 LNs for post-operative prognostic stratification for patients with LN-negative disease. On the other hand, adjuvant chemotherapy is recommended for patients with NSCLC who have any sign of LNM (17). Therefore, identification of LNM is a crucial step in management and treatment decision in patients with NSCLC.
Radiomics provides quantitative measurements of heterogeneity based on distribution of grey levels and is not affected by subjective analysis. Some studies (7,8) have shown that the predictive accuracy of CT radiomics signature was favorable for LNM in patients with colorectal cancer and bladder cancer. However, the segmentation of lesions in these studies relies on the expert’s manually contouring, which is painful for the doctor. Recent developments in deep learning provide methods performing organ and lesion segmentation. Combined automatic lesion segmentation and advanced prediction methods with radiomics provide clinicians with powerful tools. As a validation of this proposed workflow, we applied a deep learning-generated nodules mask to a radiomics pipeline in patients with primary lung ADC. Our aim was to determine whether an established automatically-generated radiomics signature could be used for the preoperative prediction of LNM.
The 3D U-net is often used for organ or lesion segmentation in medical imaging (18,19). We adapted it for nodule segmentation using four scales to cover different nodule sizes. In the LIDC datasets, the median dice coefficient among four radiologists was 0.83, whereas our 4-scale 3D U-net model achieved 0.83 in the training cohort, 0.79 in the validation cohort, and 0.80 the test cohort. Although its mask it not as good as a radiologist, it has the advantage of being automated. Less variability might accelerate the radiomics predication pipeline.
Our radiomics signature exhibited favorable discrimination, with AUCs of 0.854 and 0.803 in the training and validation cohorts, respectively. However, the diagnostic sensitivity for two cohorts was relatively low, 77.51% and 72.00%, respectively. Thereafter, we took into account CT-reported LN status, tumor size, and clinical factors to increase sensitivity of radiomics signature. A multivariate logistic regression model indicated that CT-reported LN status was a significant predictive factor, distinct from the radiomics signature. Finally, the radiomics nomogram consisting of the radiomics signature and CT-reported LN status successfully stratified patients according to their risk of LNMs, showing good calibration and discrimination in the training and validation cohorts. The AUC of the radiomics nomogram in predicting LNMs of two cohorts was 0.871 and 0.856, respectively. The diagnostic sensitivity and specificity of the two cohorts were 85.71% and 77.19%, 91.67% and 82.14%, respectively.
PET-CT is a non-invasive staging method of cancer that is increasingly employed by lung cancer multidisciplinary teams. Many studies (20-23) reviewed the diagnostic performance of PET/CT in LN staging of patients with NSCLC. A systematic review (23) showed that the summary sensitivity and specificity estimates for the activity > background PET-CT positivity criterion were 77.4% and 90.1%, respectively. However, the accuracy estimates of these studies in ROC space showed a wide prediction region, and the summary sensitivity and specificity estimates for the SUVmax ≥2.5 PET-CT positivity criterion were 81.3% and 79.4%, respectively. In these two groups, the accuracy estimates in the ROC space showed a very wide prediction region. Thus, the accuracy of PET-CT is insufficient to recommend PET-CT alone. However, the prediction of LNM by the radiomics nomogram reached or even surpassed the level of PET/CT. This may be related to the fact that microscopic LNM are barely detected by PET/CT. Recent development of radiomics enabled us to easily obtain high-throughput data without re-scanning, reducing radiation exposure. We constructed a radiomics nomogram that facilitates preoperative individualized prediction of LNM. If a radiomics nomogram is widely validated, it would require only 1–3 minutes to obtain a predicted risk of LNMs from an established model.
To justify clinical usefulness, we assessed whether the radiomics nomogram-assisted decisions would improve patient outcomes by DCA. This method offers insight into net benefits based on threshold probability. The decision curve showed that if the threshold probability of a patient or physician is between 0.46 and 0.83, using the radiomics nomogram to predict LNMs adds more benefit than either the treat-all-patients scheme or the treat-none scheme.
Some investigators (24-26) attempted to use CT texture analysis to discriminate malignant from benign LN in lung cancer. However, these studies analyzed only larger LNs and could not analyze normal-sized and occult LNM (micrometastases). A systematic review (27) suggested that micrometastases are clinically relevant in NSCLC, conferring worse prognosis. The innovation of our study is prediction of the possibility of LNM by texture analysis of the primary lesion, because the primary lesion reflects pathological features, such as tumor hypoxia and angiogenesis (28).
A valuable feature of our radiomics nomogram is its discriminatory ability in cN0 patients. Patients diagnosed as cN0 are typically considered to be at low risk for LNM. However, some cN0 patients also harbor LNMs, and it is a formidable challenge to precisely identify which patients will experience LNM. Our nomogram showed good discriminatory ability in cN0 patients to identify the patients at high risk of LNM. Furthermore, when categorized into low- and high-risk groups on the basis of the cutoff values of the risk score derived from the nomogram, the high-risk group had a significantly greater probability of having LNMs in patients with lung cancer. The high-risk group of cN0 patients will benefit by increasing the number of LN dissection intents.
There are some limitations to the present study. First, the sample size was relatively small, because we adopted standard CT data with the same machine, reconstruction algorithm and section thickness. Second, a minority of patients in our study did both CT and PET-CT before surgery because of the high cost of PET-CT and the high radiation exposure. We will add related cases in future studies and compare studies results with PET-CT. Third, we were not able to externally validate the method. Fourth, LNM in lung cancer is affected by gene expression profiles, such as those of microRNA-31 and C-Met (29,30). We did not take this into account. Fifth, we did not consider new features discovered by deep learning. At the time of this study, we considered deep learning to be a mask input to the radiomics pipeline. In the future, combined radiomics features with deep learning-discovered features might further improve the predictive accuracy automatically.
Conclusions
In conclusion, we presented a radiomics nomogram consisting of both a radiomics signature and CT-reported LN status. The nomogram showed favorable predictive accuracy for preoperative LNM and can be used for CT-reported LN-negative patients. These results may contribute to preoperative staging, prediction of treatment response and prognostic evaluation of lung cancer.
Deep learning technique (3D U-net) based nodule automatic segmentation
The process of nodule automatic segmentation consists of three steps: pre-processing, nodule localization, and nodule segmentation. For pre-processing, we first normalize to Hounsfield units in order to enhance the lung window, then the lung organ is segmented using level set and region growing techniques. For nodule localization, we introduce a multiple size scaled 3D U-net with a coverage of large range of nodules size. For nodule segmentation, the input to the segmentation 3D U-net is the bounding box proposed by localization 3D U-net. The localization 3D U-net shares the same structure as the segmentation 3D U-net, but with different input matrix size and train labels. Details on segmentation 3D U-net (Figure 1) are as follows: the input matrix to 3D U-net is 80×80×80; the proposed bounding box from localization is resampled to be adapted to the same matrix size.
The base filter size to 3D U-net 32 and increment grow size along max-pooling and deconvolution path is 32 and −32.
A detailed description of the LASSO method
LASSO is a powerful method for regression with high dimensional predictors (31). We combined the LASSO method with the Cox proportional hazard regression model for survival analysis. We used the LASSO Cox regression model to select the most important prognostic features from the training dataset. This method minimizes a log partial likelihood subject to the sum of the absolute values of the parameters being bounded by a constant:
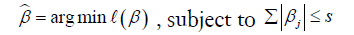
where 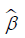 is the obtained parameters,
is the obtained parameters, 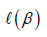 is the log partial likelihood of the Cox proportional hazard regression model, and
is the log partial likelihood of the Cox proportional hazard regression model, and 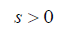 is a constant.
is a constant.
Benefiting from the absolute constraint, the LASSO method shrinks coefficients and changes some coefficients to zero. Therefore, the LASSO method can be used for feature reduction and selection. We set the standardized constraint parameter as 0.0228 and LASSO selected 14 nonzero coefficients (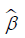 ). Then, the Cox proportional hazard regression model was obtained with its outcome h(t) being the hazard rate at time t for individuals. The “glmnet” package in R statistical software version 3.4 was used for LASSO logistic regression model analysis.
). Then, the Cox proportional hazard regression model was obtained with its outcome h(t) being the hazard rate at time t for individuals. The “glmnet” package in R statistical software version 3.4 was used for LASSO logistic regression model analysis.
Decision curve analysis (DCA)
In our study, the DCA method was used to evaluate the clinical utility of the presented nomogram. The DCA algorithm assesses prediction models by calculating the range of threshold probabilities in which a prediction or prognostic model was clinically useful (32). DCA is a compositive method for evaluating and comparing different diagnostic and prognostic models. The theory of DCA can be illustrated by the equation below:
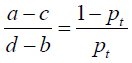
where d – b represents the influence of unnecessary treatment. If treatment is directed by a prediction model, d – b is the harm related to a false-positive result compared with a true-negative result. Inversely, a – c represents the consequence of rejecting beneficial treatment, in other words, the harm from a false-negative result compared with a true-positive result. Pt represents where the expected benefit of treatment is equal to the expected benefit of refraining from treatment.
The details of 14 selected features by using LASSO logistic regression
Acknowledgements
Funding: The research was supported by Open Project of State Key Laboratory of Respiratory Disease (SKLRD2016OP011) and Science and Technology Planning Project of Guangdong Province (Grant No. 2017A040405065).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This retrospective study was approved by the Institutional Review Board of the First Affiliated Hospital of Guangzhou Medical University [Medical Ethical Review (MER) 2017-38]. Medical record review was performed in accordance with institutional ethics review board guidelines.
References
- Jemal A, Center MM, DeSantis C, et al. Global patterns of cancer incidence and mortality rates an trends. Cancer Epidemiol Biomarkers Prev 2010;19:1893-907. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Silvestri GA, Gould MK, Margolis ML, et al. Noninvasive staging of non-small cell lung cancer: ACCP evidenced-based clinical practice guidelines (2nd edition). Chest 2007;132:178S-201S.
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [PubMed]
- Huang Y, Liu Z, He L, et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol 2018;28:582-91. [Crossref] [PubMed]
- Wu S, Zheng J, Li Y, et al. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clin Cancer Res 2017;23:6904-11. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Nambu A, Kato S, Motosugi U, et al. Thin-section CT of the mediastinum in preoperative N-staging of non-small cell lung cancer: comparison with FDG PET. Eur J Radiol 2010;73:510-7. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Med Image Comput Comput Assist Interv 2015;9351:234-41.
- Çiçek Ö, Abdulkadir A, Lienkamp SS, et al. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Med Image Comput Comput Assist Interv 2016;9901:424-32.
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Asamura H, Chansky K, Crowley J, et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the Revision of the N Descriptors in the Forthcoming 8th Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2015;10:1675-84.
- Shapiro M, Mhango G, Kates M, et al. Extent of lymph node resection does not increase perioperative morbidity and mortality after surgery for stage I lung cancer in the elderly. Eur J Surg Oncol 2012;38:516-22. [Crossref] [PubMed]
- Darling GE, Allen MS, Decker PA, et al. Randomized trial of mediastinal lymph node sampling versus complete lymphadenectomy during pulmonary resection in the patient with N0 or N1 (less than hilar) non-small cell carcinoma: results of the American College of Surgery Oncology Group Z0030 Trial. J Thorac Cardiovasc Surg 2011;141:662-70. [Crossref] [PubMed]
- Liang W, He J, Shen Y, et al. Impact of Examined Lymph Node Count on Precise Staging and Long-Term Survival of Resected Non-Small-Cell Lung Cancer: A Population Study of the US SEER Database and a Chinese Multi-Institutional Registry. J Clin Oncol 2017;35:1162-70. [Crossref] [PubMed]
- Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw 2013;11:645-53. [Crossref] [PubMed]
- Shen D, Wu G, Suk HI, et al. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Lv YL, Yuan DM, Wang K, et al. Diagnostic performance of integrated positron emission tomography/computed tomography for mediastinal lymph node staging in non-small cell lung cancer: a bivariate systematic review and meta-analysis. J Thorac Oncol 2011;6:1350-8. [Crossref] [PubMed]
- Broderick SR, Patterson GA. Performance of integrated positron emission tomography/computed tomography for mediastinal nodal staging in non-small cell lung carcinoma. Thorac Surg Clin 2013;23:193-8. [Crossref] [PubMed]
- Liao CY, Chen JH, Liang JA, et al. Meta-analysis study of lymph node staging by 18 F-FDG PET/CT scan in non-small cell lung cancer: comparison of TB and non-TB endemic regions. Eur J Radiol 2012;81:3518-23. [Crossref] [PubMed]
- Schmidt-Hansen M, Baldwin DR, Hasler E, et al. PET-CT for assessing mediastinal lymph node involvement in patients with suspected resectable non-small cell lung cancer. Cochrane Database Syst Rev 2014.CD009519. [PubMed]
- Pham TD, Watanabe Y, Higuchi M, et al. Texture Analysis and Synthesis of Malignant and Benign Mediastinal Lymph Nodes in Patients with Lung Cancer on Computed Tomography. Sci Rep 2017;7:43209. [Crossref] [PubMed]
- Andersen MB, Harders SW, Ganeshan B, et al. CT texture analysis can help differentiate between malignant and benign lymph nodes in the mediastinum in patients suspected for lung cancer. Acta Radiol 2016;57:669-76. [Crossref] [PubMed]
- Bayanati H, E, Thornhill R, Souza CA, et al. Quantitative CT texture and shape analysis: can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur Radiol 2015;25:480-7. [Crossref] [PubMed]
- Coello MC, Luketich JD, Litle VR, et al. Prognostic significance of micrometastasis in non-small-cell lung cancer. Clin Lung Cancer 2004;5:214-25. [Crossref] [PubMed]
- Ganeshan B, Goh V, Mandeville HC, et al. Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 2013;266:326-36. [Crossref] [PubMed]
- Meng W, Ye Z, Cui R, et al. MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin Cancer Res 2013;19:5423-33. [Crossref] [PubMed]
- Han CB, Ma JT, Li F, et al. EGFR and KRAS mutations and altered c-Met gene copy numbers in primary non-small cell lung cancer and associated stage N2 lymph node-metastasis. Cancer Lett 2012;314:63-72. [Crossref] [PubMed]
- Sauerbrei W, Royston P, Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med 2007;26:5512-28. [Crossref] [PubMed]
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. [Crossref] [PubMed]