The new era of whole-exome sequencing in congenital heart disease: brand-new insights into rare pathogenic variants
Prolegomena to the genetics of congenital heart disease (CHD)
CHD is a complex developmental phenotype, with many genes contributing to its etiology; it affects about 1% of infants, and its genetic roots are multifarious and difficult to ascertain (1,2). Improvements in prenatal diagnosis, corrective procedures, and longitudinal support have reduced infantile mortality. Today, >75% of CHD children surviving beyond their 1st year, including those with complex malformations, will live into adulthood (3). Despite medical and surgical advances, CHD leads birth-defect mortality. Genetic factors have long been implicated, as children with genetic disorders are more prone to heart disease.
Mutations or variants (2) underlying CHD are genomic alterations that are either inherited or appear de novo in an individual. Aside from inheriting half the genome of each parent, we are born with small numbers of such de novo mutations (DNMs) that occurred during gametogenesis and post-zygotically; nonetheless, DNMs occur as non-germline events too. Mutations that arise later in development or post-natal life can remain restricted to a single tissue or even a small number of somatic cells.
Progress in CHD management is associated with improved survival and elevated risk for delayed cardiac complications, often in a multisystem-disease context encompassing neurodevelopmental and other congenital abnormalities (4). Genetic risk denotes the odds of becoming affected by a disorder; it may be elevated because of inherited genes causing or raising susceptibility to it. Both genes and Bernardian environment (1,2,5,6) impact risk or buffer against disease and can alter phenotypic manifestations leading to different CHD subtypes. The same variants often contribute to different outcomes, depending on environmental and modifier-gene background (2,7).
Addition of “omics” to a molecular term root implies comprehensive assessment of a class of molecules (8); thus, genomics focuses on entire genomes whereas genetics on specific variants or genes. Incomplete understanding of CHD genomics renders problematic any questions about children’s future health or CHD-risk for planned offspring. But high-throughput DNA analyses, including massively-parallel next generation sequencing (NGS) (2,7,8), offer unique opportunities to discover even low-frequency or rare informative variants underlying human disease. Thus, a remarkable study appearing lately in Nature Genetics (9) updates fundamental molecular biology of cardiac development, genetic causes underlying CHD, and long-term prognosis including persisting problems risk, for individuals with such rare damaging or causal variants.
Pediatric cardiac genomics consortium (PCGC) pursues rare CHD genes
The pluri-disciplinary study, which I consider in this Editorial, examined the contribution of rare inherited and de novo variants to CHD. It was conducted by the US NHLBI’s PCGC, a part of the Bench to Bassinet Program. This Program’s mission is to accelerate scientific-discovery-to-clinical-practice by fostering an approach designed to improve outcomes for individuals with CHD while supporting pediatric heart disease research. The study utilized clinical and genetic data from 2,871 CHD probands, comprising 2,645 parent-offspring trios and 226 singletons (see below), along with information and genetic data obtained from parents (9). Large sample sizes are required to associate rare variants with complex phenotypic traits; and individual subpopulations, e.g., Ashkenazi Jews, have their own rare variants. Accordingly, patients and parents were recruited from major US Centers and University College London; exomes were sequenced at Yale University.
With the highly dense genomic data available to the PCGC, ignoring linkage between genes/loci would occasion disregard of valuable information. Accordingly, the relationship between proband and parents was estimated using pairwise identity-by-descent (IBD) calculations (10). A genome-/exome-wide IBD estimate is a metric quantifying the level of relatedness between pairs of individuals (11), useful in many applications, including identifying variants influencing disease susceptibility/risk, and heritability analysis in founder (see below) populations. Recent technological advances including NGS support sequencing the ~2% of the human genome that encompasses all coding regions, the exome (7,8), rapidly and affordably.
Whole-exome/-genome sequencing (WES/WGS) datasets from large-scale projects have been increasingly used to determine variant properties and associations (7,8). WGS locates efficaciously common variants of very small effect and WES de novo and very rare variants of larger effect. IBD was calculated using PLINK (12,13), a bioinformatics tool for WES/WGS and association studies (GWAS/EWAS) (2,7,8), and linkage analyses. With PLINK large datasets, comprising hundreds of thousands of markers genotyped for thousands of subjects, can be rapidly manipulated and analyzed. Predictably, IBD sharing between proband and parents in all trios was ~50% (45–55%).
Linking rare genetic variants to complex CHD traits
Increasing availability of NGS large-sample data has uncovered many rare mutations including “singletons,” those variants observed only in a single genome of a large sample. Explosive growth of the human population beginning ~1,400 years ago helped seed the human genome with negative variants that have not yet been discarded by natural selection (14). Rare genetic variants were not captured by association studies using single nucleotide polymorphism-chips, but with NGS technologies exploring the entire genome of individuals and not only some predefined loci where common variants have been detected they have become detectable. We can now study contributions of rare mutations to CHD, using genotyping exome-arrays encompassing rare alleles. Thousands of genetic sequences must be studied to find enough of the variants to connect them to particular CHD phenotypes. Since many of these variants are not located in protein-coding regions, sequencing exomes is insufficient to comprehensively assess genetic risk for complex CHD-syndromes (Figure 1). And it is always important to remember that both genetic mutations and environmental stressors, including pollutants, toxins, and maternal infections, may impact cardiac development (6-8), by altering gene expression in various embryonic cell-types (1,2).
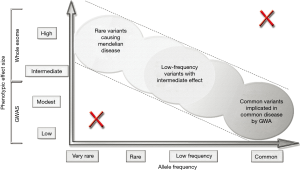
Inherited mutations
The PCGC determined that some damaging mutations are definitely transmitted from parents to children (9). Among these were mutations in the FLT4 gene that consistently led to tetralogy of Fallot (ToF) (15), a complex cyanotic CHD phenotype involving conotruncal defects. Also, mutations in the MYH6 gene that encodes MYH6 (cardiac α-myosin heavy-chain). MYH6 is highly expressed during cardiac embryogenesis; after birth, cardiac ventricles predominantly express the β-isoform, whereas atria the α-isoform (16). MYH6 variants accounted for ~11% of Shone’s complex (17), which entails left-sided cardiac malformations including supravalvular mitral membrane, parachute mitral valve, subaortic stenosis and aortic coarctation.
Some CHD patients also carried an Ashkenazi Jewish founder-mutation; the mutation in both copies of the GDF1 gene, which encodes a multifunctional cytokine of the TGF-beta (transforming growth factor-beta) superfamily of proteins (18), accounted for ~5% of serious CHD among children of Ashkenazi descent. Founder-mutations are a special class of variants embedded in identical DNA portions in all carriers. Every founder-mutation carrier has a common ancestor—the “founder”. As discrete populations mix, specific ethnicity-mutations will be encountered more randomly.
DNMs
The investigations also revealed DNMs in offspring (9), a source of genetic complexity because they arise spontaneously; the parents do not harbor the mutation and family history is negative for CHD. For highly penetrant mutations underlying severe CHD forms, this mechanism implies strong negative natural selection against the accumulation of these mutations in the population. DNMs accounted for ~8% of cases, including ~3% of isolated CHD patients and ~28% of CHD cases coexisting with neurodevelopmental and extracardiac congenital anomalies.
Such DNMs were especially frequent in genes concerned with chromatin remodeling and modifications of chromatin histone proteins—nuclear DNA is packaged into nucleosomes where the DNA double helix is wrapped around a central core of histone proteins (5-7). Nucleosomes are compact, but the ends or “tails” of the histone proteins composing them stick out from the otherwise compact nucleosome structure (7). Histone protein modifications normally phase-in processes regulating gene transcription and the epigenome, a system of chemical tags modulating gene expression.
DNMs in ~440 genes were deduced to contribute to CHD, and the investigators found a remarkable overlap of damaging DNMs implicating variants contributing to both CHD and autism (9). Most strikingly, 87% of patients who had loss-of-function (LoF) DNMs in chromatin modifiers had neurodevelopmental disorders, a shared genetic etiology that had been previously noted (19). This underscores the requirement that for a normally developed/functioning body, a sound and extensive neural network is vital. Infants with CHD tend to have weaker connections within their brain hubs; this underlies neurodevelopmental disabilities affecting motor functions, learning and cognitive processes.
Other new findings concerning rare variants
Furthermore, 12 genes that had not previously been linked to it were found to have >70% chance of association with CHD (9). Additional findings of the PCGC investigation suggested that some respiratory ailments occurring in CHD patients are linked to defects (ciliopathies) in cilia that carry out a host of biological functions; implicated genes are associated with human primary ciliary dyskinesia and situs inversus totalis. Ciliary genes are known to have a critical role in establishment of the left-right body axis, and their mutations frequently contribute to abnormal left-right relationships (heterotaxy), observed in CHD-syndromes (20).
Regulatory mechanisms of gene expression
DNA methyl tags and methyl, hydroxymethyl, acetyl and some other histone-tail tags in combination control whether an underlying gene is switched on-or-off (gene silencing). Environmental factors too can impact expression of sets of genes by altering methylation/hydroxymethylation patterns, histone modification patterns and chromatin remodeling: euchromatin is decompacted/dispersed and prevalent in cells that are active in the transcription of many of their genes while heterochromatin is compacted and most abundant in cells that are less active or not active (2,7,21).
These chemical tags do not change the DNA base sequence, but can make the DNA nearby fold or unfold (7), blocking access to a gene so it cannot turn on, or exposing a gene so it can be activated; moreover, when genes are switched-on their nucleosomes are more uncoiled. As I detailed above, multiple exogenous and endogenous environmental factors, including maternal infections and prenatal nutrition, acting on the genome during prenatal and early postnatal development, facilitate genome-wide changes in the epigenome that alter transcription at distinct developmental time-points (5,6,22). Regulated nucleosome-cluster decompaction and transcriptional activation of cardiomyocyte nuclear DNA are molecular dynamic processes enabling gene transcription leading to normal and pathologic eccentric/concentric hypertrophy (23-27).
Other regulatory mechanisms at the transcriptional level include binding of transcription factors to DNA promoter-regions, typically adjacent to the genes that they regulate. Additionally, both normal genes and variants can be regulated at the post-transcriptional level through alternative splicing, microRNA-mediated regulation, and other chemical modifications of the mRNA (28). In particular, antisense microRNA (single-stranded RNA complementary to a mRNA) can regulate gene expression by triggering sequence-specific cleavage or translation repression of the target mRNA transcripts (2,7,8,25).
Research and clinical prospects
High-throughput NGS and bioinformatics analyses enable rapid and broad sequencing of genes, entire exomes or genomes and are applicable to diagnostic assessments of CHD risk and individualized treatments (1,2,7,8,26,27). The rapid adoption of NGS-based tests in research and clinical practice is leading to identification of increasing numbers of CHD variants, including rare mutations unique to an individual or family. From their cohort-data, the PCGC researchers estimated that rare mutations contribute to ~2% of CHD cases (9); understanding pheno-genomic diversity and the clinical significance of these variants holds big future promise.
Despite GWAS/EWAS success in determining thousands of variant associations with complex cardiovascular traits, few such findings have been successfully translated into clinical CHD use (2,7,8,26,29). Phenotyping advances from exploitation of electronic health records (EHR) encompassing solid-state multisensor high-fidelity catheterization signs (1,2,5,7,8,23-27,30) should further boost the power of current GWAS/EWAS approaches (Figure 2). In translating multimodal lab-results to clinical practice, the PCGC findings (9) should be advantageous in expanding genetic testing panels for CHD and furnishing information for parents about recurrence risks in planned children.
As with any evolving scientific subject, our grasp of personal genomics, genomic variants, and CHD risks and cures should escalate in the upcoming years. Near term, we anticipate continued improvements in genomic sequencing-technology; enhanced understanding of normal and abnormal heart-development and its relation to broader (patho-)physiology, improving management of common and rare variants; perfected bioinformatics methodologies and software for applying CHD genomics effectively in the clinic; and upgraded Information-Technology infrastructure, tools and societal support, for sharing and analyzing genomic materials using the Internet-Cloud.
Personalized medicine implications for CHD
Revealing variants underlying individual patients’ CHD can guide direct corrective approaches to phenotypic anomalies, optimizing outcomes. Since large numbers of genes/loci may be contributing to CHD, WES/WGS would represent the all-embracing information-gathering option, preferable to looking out for a particular candidate-gene mutation. Aggregation of clinical genotype-phenotype linkages into genetic variant databases and substantiation for these associations following well-defined guidelines will expedite translation of causative variant information into clinical insights and treatments. Genetic variant databases should clearly document evidence-sources (e.g., literature, well-documented case histories, etc.) supporting variant effect interpretations.
Hopefully these advances, amplifying the pioneering work of the PCGC (9), will promote a personalized CHD medicine. To achieve this, a distinctive relationship is required between patients, cardiologists, genomic investigators, and the smart digital information and diagnostics industries. As genomic cardiology becomes more accurate and affordable, a new paradigm is emerging, in which diagnostics nurtures breakthrough therapies (2,7,8). Pertinent current diagnostics advances encompass: cell-free fetal DNA testing, using a blood sample from an expectant mother to analyze fetal DNA leaked into her bloodstream, for CHD variant(s); liquid biopsy system to detect biomarker gradients in coronary arteries giving information on atherosclerotic disease; and testing for various analytes in exhaled breath-analysis.
Conclusions and future directions
Translational research today needs to focus on the individual CHD-patient with clinical and multimodal-diagnostic phenotyping linked to discovery-platforms encompassing EHR and inexpensive cloud data-processing, and capable of uncovering causative genomic/proteomic pathways. By applying systems methodologies, it should be possible to identify a few highly-connected biochemical nodes (hubs) where causal pathways linked to specific CHD (sub-)phenotypes converge; such hubs are likely to be critical targets for various interventions.
Innovative in vitro pathway-based human and animal models should validate treatment effects on identified targets. These endeavors will require novel bioinformatics approaches to phenotyping (e.g., machine-learning algorithms allowing computers to learn-and-infer from data), and production of simulated disease in virtual patients to enable greater accuracy in predicting CHD outcomes with-and-without interventions during prenatal cardiac development and in postpartum life.
Because of past rapid population growth with inadequate purifying natural selection, human populations harbor an abundance of rare variants, many of which have relevance to understanding CHD risk. The current rapid human population growth likewise exacerbates the load of rare pathogenic variants and can contribute to the individual genetic burden of CHD; consequently, this extreme population growth merits attention in studying the genetics of complex CHD phenotypic traits.
Although identifying rare inherited and DNMs is becoming increasingly easy, interpreting them (i.e., linking them to a disease phenotype) often remains challenging. Pinpointing the genetic cause of CHD-syndromes caused by DNMs can be challenging clinically, because of pleiotropy and genetic heterogeneity underlying a single phenotype. In coming years, we will experience continued growth in numbers of rare genes identified, and deepening comprehension of the causal genetic architecture of CHD. Before turning this knowledge into syndrome classification based on underlying genotype and into widespread genomic CHD-treatments, function needs to be assigned to thousands of small-effect variants. The thought-provoking PCGC study appearing recently in Nature Genetics presages speedy progress along these objectives.
Acknowledgements
Funding: Research support, for work from my Laboratory surveyed here, was provided by: National Heart, Lung, and Blood Institute, Grant R01 HL 050446; National Science Foundation, Grant CDR 8622201; and North Carolina Supercomputing Center and Cray Research.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Pasipoularides A. Heart's Vortex: Intracardiac Blood Flow Phenomena. Shelton: PMPH USA, Ltd., 2010.
- Pasipoularides A. Linking genes to cardiovascular diseases: Gene action and gene-environment interactions. J Cardiovasc Transl Res 2015;8:506-27. [Crossref] [PubMed]
- Gilboa SM, Salemi JL, Nembhard WN, et al. Mortality resulting from congenital heart disease among children and adults in the United States, 1999 to 2006. Circulation 2010;122:2254-63. [Crossref] [PubMed]
- Bagge CN, Henderson VW, Laursen HB, et al. Risk of dementia in adults with congenital heart disease: Population-based cohort study. Circulation 2018;137:1912-20. [Crossref] [PubMed]
- Pasipoularides A. Mechanotransduction mechanisms for intraventricular diastolic vortex forces and myocardial deformations: Part 1. J Cardiovasc Transl Res 2015;8:76-87. [Crossref] [PubMed]
- Pasipoularides A. Mechanotransduction mechanisms for intraventricular diastolic vortex forces and myocardial deformations: Part 2. J Cardiovasc Transl Res 2015;8:293-318. [Crossref] [PubMed]
- Pasipoularides A. Implementing genome-driven personalized cardiology in clinical practice. J Mol Cell Cardiol 2018;115:142-57. [Crossref] [PubMed]
- Pasipoularides A. Genomic translational research: Paving the way to individualized cardiac functional analyses and personalized cardiology. Int J Cardiol 2017;230:384-401. [Crossref] [PubMed]
- Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature Genetics 2017;49:1593. [Crossref] [PubMed]
- Browning SR. Estimation of pairwise identity by descent from dense genetic marker data in a population sample of haplotypes. Genetics 2008;178:2123-32. [Crossref] [PubMed]
- Day-Williams Aaron G, Blangero J, Dyer Thomas D, et al. Linkage analysis without defined pedigrees. Genet Epidemiol 2011;35:360-70. [Crossref] [PubMed]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-75. [Crossref] [PubMed]
- Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [Crossref] [PubMed]
- Gao F, Keinan A. Explosive genetic evidence for explosive human population growth. Curr Opin Genet Dev 2016;41:130-9. [Crossref] [PubMed]
- Lillehei CW, Varco RL, Cohen M, et al. The first open heart corrections of tetralogy of Fallot. A 26-31 year follow-up of 106 patients. Annals of Surgery 1986;204:490-502. [Crossref] [PubMed]
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 1996;76:371-423. [Crossref] [PubMed]
- Shone JD, Sellers RD, Anderson RC, et al. The developmental complex of "parachute mitral valve," supravalvular ring of left atrium, subaortic stenosis, and coarctation of aorta. Am J Cardiol 1963;11:714-25. [Crossref] [PubMed]
- Herpin A, Lelong C, Favrel P. Transforming growth factor-β-related proteins: an ancestral and widespread superfamily of cytokines in metazoans. Dev Comp Immunol 2004;28:461-85. [Crossref] [PubMed]
- Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015;350:1262-6. [Crossref] [PubMed]
- Ware SM, Aygun MG, Hildebrandt F. Spectrum of clinical diseases caused by disorders of primary cilia. Proc Am Thorac Soc 2011;8:444-50. [Crossref] [PubMed]
- Pasipoularides A. Diastolic filling vortex forces and cardiac adaptations: probing the epigenetic nexus. Hellenic J Cardiol 2012;53:458-69. [PubMed]
- Grayson DR, Guidotti A. Merging data from genetic and epigenetic approaches to better understand autistic spectrum disorder. Epigenomics 2016;8:85-104. [Crossref] [PubMed]
- Pasipoularides A. Clinical assessment of ventricular ejection dynamics with and without outflow obstruction. J Am Coll Cardiol 1990;15:859-82. [Crossref] [PubMed]
- Pasipoularides A. Calcific aortic valve disease: Part 1 Molecular pathogenetic aspects, hemodynamics, and adaptive feedbacks. J Cardiovasc Transl Res 2016;9:102-18. [Crossref] [PubMed]
- Pasipoularides A. Calcific aortic valve disease: Part 2 Morphomechanical abnormalities, gene reexpression, and gender effects on ventricular hypertrophy and its reversibility. J Cardiovasc Transl Res 2016;9:374-99. [Crossref] [PubMed]
- Pasipoularides A. Know me! Unraveling the riddle of calcific aortic valve disease by bioinformatics. Tohoku J Exp Med 2017;243:255-61. [Crossref] [PubMed]
- Pasipoularides A. Challenges and controversies in hypertrophic cardiomyopathy: clinical, genomic and basic science perspectives. Rev Esp Cardiol (Engl Ed) 2018;71:132-8. [Crossref] [PubMed]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 2007;8:93-103. [Crossref] [PubMed]
- Allard D, Amsellem S, Abifadel M, et al. Novel mutations of the PCSK9 gene cause variable phenotype of autosomal dominant hypercholesterolemia. Hum Mutat 2005;26:497. [Crossref] [PubMed]
- Pasipoularides A. LV twisting and untwisting in HCM: ejection begets filling. Diastolic functional aspects of HCM. Am Heart J 2011;162:798-810. [Crossref] [PubMed]


