The correlation between crizotinib efficacy and molecular heterogeneity by next-generation sequencing in non-small cell lung cancer
Introduction
Lung cancer is the primary reason for malignant tumour morbidity and death globally and in China. Non-small cell lung cancer (NSCLC) accounts for 80–85% of all lung cancer patients. Patients usually lose the opportunity for surgery when they first visit the doctor due to the high malignancy of lung cancer, which leads to the inclination towards conservative treatment (1,2). In recent years, with the rapid progression of molecular level investigations in lung cancer, several driver mutations have been recognized, including the epidermal growth factor receptor (EGFR) gene and the EML4-ALK rearrangement, and the treatment therapy of NSCLC has been remarkably changed. The therapeutic regimens based on the targeted tyrosine kinase have gained rapid and striking therapeutic achievements in specific genotype populations (3-6).
In 2007, Soda et al. (7) first demonstrated lung adenocarcinoma-associated gene fusion in resected lung adenocarcinoma specimens, the echinoderm microtubule-associated protein-like-4-anaplastic lymphoma kinase (EML4-ALK), which contains exons 1 to 13 of EML4 and exons 20 to 29 of ALK. EML4-ALK is carcinogenic in vivo and in vitro. Similar to the carcinogenesis mechanism induced by EGFR mutations, the development of tumours is manifested as a dependency on oncogenes. EML4-ALK fusion-driven cancers accounted for 5% of NSCLC (8) and were called “ALK-positive NSCLC” (9). In NSCLC patients, a variety of EML4-ALK fusion variants have been identified that contain the same ALK but different EML4 portions (9-11). The most common variants are variant 1 and variant 3, these two variants accounts for 33% and 29% of ALK-positive NSCLC, respectively (12-14). Furthermore, nearly all of the EML4-ALK fusion variants are carcinogenic and ALK-dependent according to previous studies (15,16).
Four methods have been developed to identify the presence of the EML4-ALK rearrangement in clinical specimens. Fluorescence in situ hybridization (FISH) and immunohistochemistry (Ventana IHC) are commonly used to detect ALK rearrangement of NSCLC (17-19). In addition, real-time polymerase chain reaction (RT-PCR) has been shown to adequately identify the presence of ALK fusion oncogenes. However, because of its high complexity and demand for specimens, the use of RT-PCR as a diagnostic tool is not currently recommended (15,20,21). Novel next-generation sequencing (NGS) techniques have been shown to accurately predict the presence of ALK rearrangements compared with other methods (22).
Crizotinib was the first tyrosine kinase inhibitor (TKI) approved by the FDA for the efficacious treatment of ALK fusion-positive NSCLC, opening up a new era of target therapy for ALK-positive NSCLC patients. However, treatment with crizotinib will inevitably lead to drug resistance, which often occurs after 1–2 years [median progression-free survival (mPFS): 10 months] of treatment via various mechanisms (23,24). Lung cancer has heterogeneous features. Previous studies have reported that there is tumour heterogeneity in different lung cancer patients with EGFR mutations and in different parts of the same patient, which affects the response to EGFR-TKIs (25,26). Hou et al. (27) also found a high concordance of ALK rearrangement between the primary tumour and paired metastatic lymph nodes in patients with lung adenocarcinoma. However, the association of tumour heterogeneity in different ALK-positive patients and the response to ALK-TKIs remain unclear. Several studies have revealed that ALK variant types had different responses to crizotinib, but these results were not concordant (28-32). In this report, we retrospectively studied the relationship between the efficacy of crizotinib and the gene mutation profile, ALK fusion type, and AF in ALK-positive NSCLC patients.
Methods
Patients and selections
Patients who met the following criteria were enrolled from May 2013 to June 2016: patients with advanced ALK-positive NSCLC treated with crizotinib in three institutions. We mainly counted the PFS of patients taking crizotinib. The study design was approved by the ethics committee of Nanjing General Hospital, who waived the need for informed consent because of the non-invasive nature of the study and patient anonymity.
Tissue DNA extraction
DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA concentration was measured using the Qubit dsDNA assay.
NGS library preparation
DNA shearing was performed with a Covaris M220, followed by end repair, phosphorylation and adaptor ligation. Fragments with the size of 200–400 bp were selected using Agencourt AMPure bead (Beckman Coulter, California, USA). Samples were hybridized with capture probes baits, followed by hybrid selection with magnetic beads and PCR amplification. The quality and size of the fragments were evaluated by a high-sensitivity DNA assay. Indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., California, USA) with pair-end reads.
Sequence data analysis
Sequence data were mapped to the human genome using BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan. Variants were filtered using VarScan fpfilter pipeline. According to the ExAC, 1000 Genomes, dbSNP, ESP6500SI-V2 database, variants with population frequency over 0.1% were grouped as single nucleotide polymorphism (SNP) and excluded from further analysis. The remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translation analysis was performed using both Tophat2 and Factera 1.4.3.
Efficacy evaluation
Patients were treated with crizotinib at a dosage of 250 mg, twice a day, and all patients were routinely taking the drug until progression of disease (PD) or unbearable side effects. Patients were regularly reviewed by computed tomography (CT) according to their medical advice. The best tumour response was evaluated according to the CT assessment using RECIST 1.1 (33). PFS was defined as the time period from medication to disease progression.
Statistical analysis
Statistical analysis was performed using SPSS version 21.0 (IBM, Armonk, NY, USA). The different clinical characteristics between groups were compared using an independent samples test and chi-square test (or Fisher’s exact test). PFS was measured from the start of crizotinib administration until the date of PD according RECIST. The survival probabilities were estimated using the Kaplan-Meier method, where differences in the variables were calculated using the log-rank test. All P values were two-sided, and P<0.05 was considered significant.
Results
Patient characteristics
From May 2013 to June 2016, 56 tissue biopsies from 52 patients with advanced NSCLC were enrolled in this study, and four patients failed DNA extraction and were removed from the group (Figure 1). The median age of the cohort was 50.8 years old, ranging from 27.0 to 77.0. Of this patient group, 48 patients had detailed records of their clinical information. Eighteen (37.5%) patients were male, and 30 (62.5%) patients were female. Fifteen (31.3%) patients had a history of smoking, and 33 (68.8%) were never smokers. Thirty-nine (81.3%) patients were diagnosed with adenocarcinoma, 2 (4.2%) with squamous carcinoma, 6 (12.5%) with adenosquamous carcinoma and 1 (2.1%) with sarcomatoid carcinoma. In addition, 35 (72.9%) patients had IIIB and IV stage disease when first diagnosed, and 13 patients had relapsed after surgery. Crizotinib was administered to 48 patients. Twenty-one (43.8%) patients received the drug as a first-line treatment of advanced or recurrent disease, 16 (33.3%) patients received it as a second-line treatment, 7 (14.6%) patients received it as a third-line or later treatment, and four patients had no related clinical information. Finally, 11 patients had brain metastasis, and 20 patients had bone metastasis (Table S1).
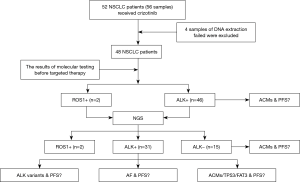

Full table
Mutation spectrum
To profile the mutation associated with each patient, we performed ultra-deep targeted sequencing using the OncoScreen panel, which can identify gene aberrance of 287 critical exons and 22 introns associated with lung cancer, with an average detection depth of 1,000×. We identified mutations from 50 (89.3%) samples, and the other 6 (10.7%) samples had no detected mutation in this panel. Several key genetic aberrances associated with lung cancer were listed in the mutation spectrum for analysing the detailed pattern of these mutations (Figure 2A).
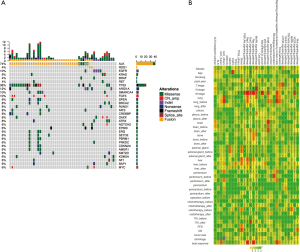
ALK was the most frequently mutated gene, accounting for 72.0% (36 ALK-mutated samples from 33 patients) of all 50 variant-containing samples identified. Thirty-six samples were detected with ALK fusion, and two samples had an ALK single-site mutation. Two (4.0%) samples were detected with ROS1 fusion. EGFR exon 19 deletion was found in 3 (6.0%) biopsies, whereas a L858R mutation was also detected in 1 (2.0%) sample. For other single-site missense mutations of oncogenic driver genes, 3 (6.0%) samples carried KRAS G12X and 2 (4.0%) samples had a BRAF V600E mutation (Figure 2A). Additionally, additional coexisting mutations (ACMs) were detected by NGS in view of tumour heterogeneity. In the 36 patients, the other coexisting mutant genes and frequencies were TP53 (n=11, 30.6%), FAT3 (n=6, 16.7%), SPEN (n=5, 13.9%), RUNX1 (n=5, 13.9%), SMARCA4 (n=4, 11.1%), BRCA2 (n=3, 8.3%), SETD2 (n=3, 8.3%), EGFR 19del (n=2, 5.6%), EGFR L858R (n=1, 2.8%), and BRAF V600E (n=1, 2.8%) (Figure 2A). The highest of mutation frequency was TP53 (30%), followed by FAT3 (17%), in ALK-positive NSCLC.
Our data revealed that metastatic stage was strongly associated with ErbB, MAPK, Ras, FoxO, Rap1, and the PI3K/Akt signalling pathway. Brain metastasis was significantly correlated with the ErbB signalling pathway (P<0.05). Best response (BR) seems to have a close relationship with ALK fusion and the pathways related to Ras, Rap1 and the pluripotency of stem cells (P<0.05) (Figure 2B).
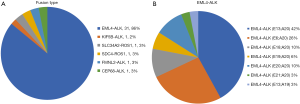
Through NGS, we also discovered several fusion gene types (Figure S1A). EML4-ALK fusion (31 samples), which consisted of several different fusion patterns, became the main ALK fusion type. Among all the EML4-ALK fusions, variant 1 (E13:A20) accounted for 42% of EML4-ALK fusion samples, whereas variant 2 (E20:A20) and variant 3 (E6:A20) represented 10% and 26%, respectively (Figure S1B). This result was consistent with previous findings (12-14).
Relationship analysis of positive ALK fusion detection by NGS and PFS of patients
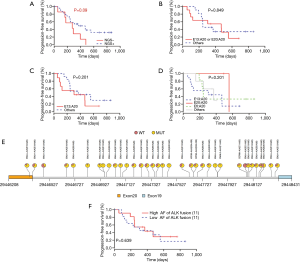
Before the ultra-deep targeted sequencing by the 295-gene panel, the cohort of 52 NSCLC patients (56 tissue samples) was detected for ALK fusion by IHC, and 46 samples were found to be positive. However, by NGS technology, only 31 samples were detected to be carrying ALK fusion. We divided the 46 ALK fusion-positive samples identified by IHC into the two following groups: NGS+ and NGS−. We further analysed the correlation of PFS after crizotinib treatment and the ALK fusion detection result by NGS. Kaplan-Meier plotting revealed that the ALK fusion patients detected as IHC+/NGS− were commonly associated with a shorter PFS compared with the ALK fusion IHC+/NGS+ patients (274 vs. 370 days, P=0.090) (Figure 3A). We speculated that the difference in time between the two detection methods might be caused by tumour heterogeneity or might be related to the inconsistency of gene transcription and protein expression.
Association of ALK variants and allelic fraction on clinical efficacy of crizotinib
We further analysed the relationship of EML4-ALK fusion type with PFS. When we combined variant 1 and variant 2 as one group, the median PFS was slightly longer than other EML4-ALK fusion types (469 vs. 246 days, P=0.849) (Figure 3B). No apparent difference could be observed among variant 1, variant 3 and the other uncommon variants (v1 =305 days, v3 =242 days, other 3 =370 days) (Figure 3C,D). Furthermore, it appeared that the PFS of patients harbouring variant 2 ALK fusion (PFS =557 days) was longer than that of the other three groups despite the limited sample quantity. Although we are unable to make definite conclusions regarding the relationship of variant type and PFS from these results due to the small group of samples and the unreliable P value, this study still provided real-world clinical evidence for further investigation of the correlation between ALK fusion type and PFS.
Next, the effect of ALK fusion variant AF on the efficacy of crizotinib was investigated. As shown in Figure 3E, it was not difficult to determine that almost every patient’s ALK fusion abundance was different. We detected the AF of ALK fusion variants in different patients by the ultra-deep targeted sequencing, and no statistically significant difference existed in the relationship between AF and PFS (P=0.639) (Figure 3F). It was suggested that AF did not affect the prognosis of patients.
Clinical outcomes with variant 1/2 and others
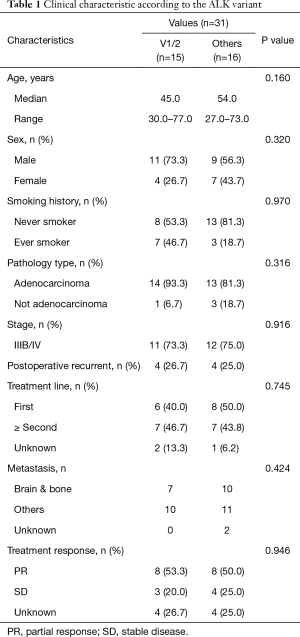
Table 1 lists the correlation between the patient’s clinical characteristics and the ALK fusion variant type. As described above, 31 patients had ALK fusion, and the presence of fusion variants was detected. Comparable clinical characteristics were observed in the variant 1/2 and other groups. We analysed the effects of different variants on the clinical characteristics of patients and found that no P values were significant. Therefore, we can conclude that variants do not affect the distribution of patient-related clinical features (age, sex, smoking history, stage, etc.) (Table 1).
Full table
The relationship between ACMs and PFS
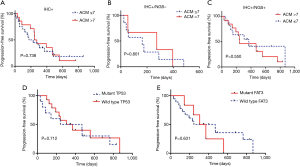
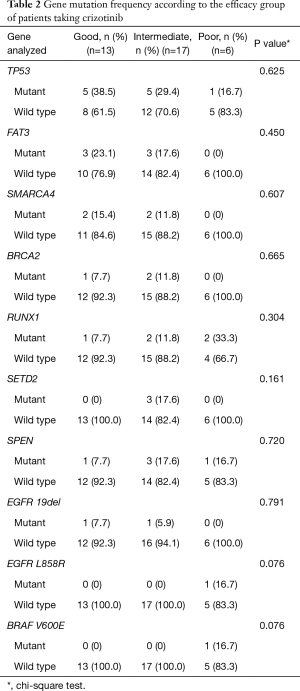
In this study, we identified 30 ACMs by NGS (Figure 2A). We divided 46 patients into three groups (IHC+, n=46; IHC+/NGS−, n=15; IHC+/NGS+, n=31) and explored the correlation between the three groups of ACMs and PFS; no significant difference (P=0.738, 0.801, 0.550) existed among these groups (Figure 4A,B,C). We further analysed the relationship between TP53, FAT3 mutation and PFS, and the results showed no significant difference (P=0.712, 0.631) (Figure 4D,E). Due to the lack of PFS in some patients, we finally selected 36 patients and divided them into three groups with good (n=13), intermediate (n=17) and poor (n=6) responders (Table S2), according to the treatment resistance to crizotinib and PFS. The median PFS was 1.16, 7.16 and 19.51 months for poor, intermediate and good responders, respectively. We assessed the correlation of the three groups and ACMs and found no significant difference (Table 2). Therefore, we could speculate that the presence of ACMs does not affect the response of ALK-positive NSCLC patients to crizotinib.

Full table
Full table
Discussion
In recent years, increasing studies have focused on the consistency of ALK-positive detection methods. Marchetti et al. (34) compared the sensitivity of RT-PCR/IHC/FISH/NGS and found that the ALK fusion detection rates of these methods were different. Ma et al. (35) enrolled six patients with IHC-positive ALK fusion, who were FISH-negative and were further examined by qRT-PCR/NGS and other methods, and only two patients had ALK fusion detected by NGS. These six patients with ALK inhibitors had a certain effect. Therefore, it is essential to further increase the consistency between the different detection methods. NGS is a promising method for the detection of ALK fusion and can capture all the breakpoint variants of ALK fusion genes. In this study, we used an NGS-based method in a cohort of 56 tissue samples from 52 patients to perform ultra-deep targeted sequencing. From the 46 samples found to be ALK-positive by IHC, NGS detected only 31 biopsies carrying positive ALK fusion. The difference between these two methods can be caused by several reasons. One possibility is the degradation of DNA that led to the false negative with NGS detection. Another reason could be explained as the real situation that no DNA mutation occurred and that the highly expressed protein level resulted in the false positive of IHC. A FISH test could be carried out to help understand ALK protein expression, which is independent on the DNA alteration. Unfortunately, the FISH method was not used for verification in this study. Our study was the first to compare the differences in the PFS between IHC+/NGS+ and IHC+/NGS− group of patients, but the specific mechanism we did not carry out in-depth exploration. Since we did not compare the PFS between chemotherapy patients and NGS patients, we do not know how much benefit NGS patients take with crizotinib. In future studies, multiple detection methods could be considered together to increase the understanding of EML4-ALK.
Previous studies (28-31,36) reported that the difference of ALK fusion variant type has a significant effect on the efficacy of crizotinib therapy for patients with ALK mutant NSCLC, although the results are not completely consistent. Heuckmann et al. (28) found that different types of ALK fusion variants also affect the efficacy of ALK-TKIs. The explanation can be illustrated by the fact that ALK variants were different in terms of their stability, so they displayed different responses to ALK inhibitors, depending on the N-terminus of EML4-ALK. Furthermore, the protein formed by variant 2 is the most unstable type and had the strongest response to ALK inhibitors. However, their experiments are only limited to in vitro, and further studies were needed to convert the studies into in vivo and clinical. Professor Yilong Wu (31) found that the fusion variant had no effect on PFS of advanced NSCLC patients, whereas the ratio of ALK-positive cells was weakly correlated with prognosis. In a study of Koreans (29), 54 ALK-positive NSCLC patients were enrolled to examine the relationship of ALK fusion variant type and the efficacy of crizotinib. They found that variant 3 showed the strongest resistance compared to variants 1/2/others. Similarly, in another study reported by Japanese researchers in 2016 (30), variant 1 had the strongest susceptibility to crizotinib. In our study, ALK fusion types were divided into two groups (variants 1/2 and others uncommon variants) based on protein stability differences (28). We found that the former variant group (variant 1 + variant 2) displayed a longer PFS trend compared to the other uncommon variants. However, no significant difference could be observed among variant 1, variant 3, and the other variants. It appeared that variant 2 had a greater impact on the patient’s PFS than the other three variant groups. Just as Heuckmann et al. (28) demonstrated, protein instability was a reason that the different types of ALK fusion variants affected the efficacy of ALK-TKIs. Although definite conclusions could not be made due to the small group, this study could also provide clinical evidence for the relationship between PFS and ALK fusion variant types. Investigations with a larger population will be needed for further study. At present, there is little research on the effect of ALK fusion variant allele fractions on the efficacy of crizotinib. Bellini et al. (32) reported the ALK-mutated allele fraction was related to the aggressive of neuroblastoma and could affect the efficacy of targeted drugs. Based on the above ideas, we detected the abundance of ALK fusion variants in different patients by NGS and found that there was no difference between AF and PFS.
Lung cancer is one of the cancers that is most prone to distant metastasis, and common metastatic sites include the brain, bone, liver, and adrenal glands. In NSCLC, the overexpression and activation of these mutual traffic signal transduction pathways are closely related to the occurrence and development of tumours. Ming et al. (37) reported that the activation of ERK1/2 could promote the expression of vascular endothelial growth factor (VEGF), cause angiogenesis, and promote distant metastasis of lung cancer. In addition, gene mutations in the PI3K-AKT-mTOR signalling pathway also contribute to the development of brain metastases (38). In this study, we found that the metastatic stage was related with ErbB, MAPK, Ras, FoxO, Rap1, and the PI3K/Akt signalling pathway. The above results suggest that the mechanism of organ metastasis of NSCLC is very complex and involves a variety of signal pathway interactions. There are great future research prospects.
Bria et al. (26) reported that ACMs affect the response to gefitinib in EGFR mutant NSCLC patients. They found that ACMs and TP53 mutations were negatively correlated with the efficacy of gefitinib. However, in this study, we did not find that ACMs affected the reactivity of patients with ALK-positive NSCLC patients for crizotinib. It is necessary to expand the sample size to further explore this topic.
There are several limitations in this study. First, the small cohort of patients resulted in uncertain conclusions regarding the correlation of ALK variant types and PFS, which were not statistically significant, although a trend could be seen. Furthermore, the obtained tissue biopsies could not represent the overall gene aberrations due to the tumour heterogeneity, which would impact the tumour response and sensitivity to crizotinib.
Conclusions
Our study is the first report to investigate the relevance of ALK+ by NGS and the clinical outcome in NSCLC. Suggesting that a variety of detection methods could be used in combination to better understand ALK, and the correlation between the different contents of heterogeneity of ALK and TKI effect is a promising research area.
Acknowledgements
We thank all study participants of the Department of Pathology for their contributions to this project and the Burning Rock Biotech which did the targeted next-generation sequencing. We apologize to all researchers whose relevant contributions were not cited due to space limitations.
Funding: This work was supported by Clinical Science and Technology Project of Jiangsu Province (No. BL2013026), the National Natural Science Foundation of China (No. 81401903), the National Natural Science Foundation of China (No. 81572273), the National Natural Science Foundation of China (No. 81602015), the Natural Science Foundation of Jiangsu Province (No. BK20161386), the National Natural Science Foundation of China (No. 81602015) and Pfizer Investigator Funding.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study design was approved by the ethics committee of Nanjing General Hospital (No. 2015NZKY-019-01), who waived the need for informed consent because of the non-invasive nature of the study and patient anonymity.
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin 2008;58:71-96. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol 2009;4:1450-4. [Crossref] [PubMed]
- Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008;14:4275-83. [Crossref] [PubMed]
- Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res 2008;68:4971-6. [Crossref] [PubMed]
- Ou SH, Bartlett CH, Mino-Kenudson M, et al. Crizotinib for the treatment of ALK-rearranged non-small cell lung cancer: a success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist 2012;17:1351-75. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Sasaki T, Rodig SJ, Chirieac LR, et al. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer 2010;46:1773-80. [Crossref] [PubMed]
- Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin Oncol 2009;27:4232-5. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008;14:6618-24. [Crossref] [PubMed]
- Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A 2008;105:19893-7. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Akerley W, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. J Natl Compr Canc Netw 2016;14:255-64. [Crossref] [PubMed]
- Conklin CM, Craddock KJ, Have C, et al. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013;8:45-51. [Crossref] [PubMed]
- Ali G, Proietti A, Pelliccioni S, et al. ALK rearrangement in a large series of consecutive non-small cell lung cancers: comparison between a new immunohistochemical approach and fluorescence in situ hybridization for the screening of patients eligible for crizotinib treatment. Arch Pathol Lab Med 2014;138:1449-58. [Crossref] [PubMed]
- Cruz-Rico G, Aviles-Salas A, Segura-Gonzalez M, et al. Diagnosis of EML4-ALK Translocation With FISH, Immunohistochemistry, and Real-time Polymerase Chain Reaction in Patients With Non-Small Cell Lung Cancer. Am J Clin Oncol 2017;40:631-8. [Crossref] [PubMed]
- Takeuchi K, Togashi Y, Kamihara Y, et al. Prospective and clinical validation of ALK immunohistochemistry: results from the phase I/II study of alectinib for ALK-positive lung cancer (AF-001JP study). Ann Oncol 2016;27:185-92. [Crossref] [PubMed]
- Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L, et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist 2015;20:316-22. [Crossref] [PubMed]
- Lin YT, Yu CJ, Yang JC, et al. Anaplastic Lymphoma Kinase (ALK) Kinase Domain Mutation Following ALK Inhibitor(s) Failure in Advanced ALK Positive Non-Small-Cell Lung Cancer: Analysis and Literature Review. Clin Lung Cancer 2016;17:e77-94. [Crossref] [PubMed]
- Matikas A, Kentepozidis N, Georgoulias V, et al. Management of Resistance to Crizotinib in Anaplastic Lymphoma Kinase-Positive Non-Small-cell Lung Cancer. Clin Lung Cancer 2016;17:474-82. [Crossref] [PubMed]
- Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol 2009;20:696-702. [Crossref] [PubMed]
- Bria E, Pilotto S, Amato E, et al. Molecular heterogeneity assessment by next-generation sequencing and response to gefitinib of EGFR mutant advanced lung adenocarcinoma. Oncotarget 2015;6:12783-95. [Crossref] [PubMed]
- Hou L, Ren S, Su B, et al. High concordance of ALK rearrangement between primary tumor and paired metastatic lymph node in patients with lung adenocarcinoma. J Thorac Dis 2016;8:1103-11. [Crossref] [PubMed]
- Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 2012;18:4682-90. [Crossref] [PubMed]
- Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791-7. [PubMed]
- Yoshida T, Oya Y, Tanaka K, et al. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383-9. [Crossref] [PubMed]
- Lei YY, Yang JJ, Zhang XC, et al. Anaplastic Lymphoma Kinase Variants and the Percentage of ALK-Positive Tumor Cells and the Efficacy of Crizotinib in Advanced NSCLC. Clin Lung Cancer 2016;17:223-31. [Crossref] [PubMed]
- Bellini A, Bernard V, Leroy Q, et al. Deep Sequencing Reveals Occurrence of Subclonal ALK Mutations in Neuroblastoma at Diagnosis. Clin Cancer Res 2015;21:4913-21. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Marchetti A, Pace MV, Di Lorito A, et al. Validation of a new algorithm for a quick and easy RT-PCR-based ALK test in a large series of lung adenocarcinomas: Comparison with FISH, immunohistochemistry and next generation sequencing assays. Lung Cancer 2016;99:11-6. [Crossref] [PubMed]
- Ma D, Wang Z, Yang L, et al. Responses to crizotinib in patients with ALK-positive lung adenocarcinoma who tested immunohistochemistry (IHC)-positive and fluorescence in situ hybridization (FISH)-negative. Oncotarget 2016;7:64410-20. [PubMed]
- Cha YJ, Kim HR, Shim HS. Clinical outcomes in ALK-rearranged lung adenocarcinomas according to ALK fusion variants. J Transl Med 2016;14:296. [Crossref] [PubMed]
- Ming J, Liu N, Gu Y, et al. PRL-3 facilitates angiogenesis and metastasis by increasing ERK phosphorylation and up-regulating the levels and activities of Rho-A/C in lung cancer. Pathology 2009;41:118-26. [Crossref] [PubMed]
- Li Q, Wu H, Chen B, et al. SNPs in the TGF-beta signaling pathway are associated with increased risk of brain metastasis in patients with non-small-cell lung cancer. PLoS One 2012;7:e51713. [Crossref] [PubMed]


