miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is the most frequently diagnosed cancer and the most common cause of cancer death worldwide, accounting for an estimated 1.8 million new cases and 1.6 million deaths in 2012 (1). Approximately 85% of lung cancers are classified as non-small cell lung cancer (NSCLC), a less aggressive form of tumor than small cell lung cancer (the remaining 15% of all lung cancers). According to Surveillance, Epidemiology, and End Results (SEER) Cancer Statistics Review, the 5-year survival rate of NSCLC remains as low as 23.6% because only 19% of cases are diagnosed at localized stages when the disease is highly curable with a 59.5% 5-year survival rate (2).
An early detection of lung cancer therefore is able to reduce the mortality to a large extent. Low-dose computed tomography (LDCT) is recommended as standard screening for the high-risk population, considering a lung cancer mortality benefit of 20% demonstrated by the National Lung Screen Trial (NLST) (3). However, a high false positive rate of 96% was shown in the NLST and could give rise to increasing screening rounds, frequent radiation exposure and exaggerated anxiety, although those results were eventually proved to be benign nodules. To complement the overdiagnosis by LDCT, specific cancer biomarkers, particularly miRNAs, would contribute to a favorable laboratory examination as discussed by numerous studies (4,5).
MicroRNAs (miRNAs) are endogenous small non-coding RNAs of 18–24 nucleotides in length, acting as important regulators in post-transcriptional gene expression. More than 2,500 mature human miRNAs have been identified so far, according to the miRBase database (http://www.mirbase.org/, version 21, accessed January 2018) since the first human-encoded miRNA, let-7, was discovered in 2000. Owing to their distinct characteristics, miRNAs have been extensively researched for the past decade for their role as potential biomarkers and novel therapeutics (6,7).
miRNA biogenesis and function
Initially, microRNA genes are transcribed by RNA polymerase II into primary miRNAs (pri-miRNAs) in the nucleus. The pri-miRNAs are subsequently cleaved by Drosha/DGCR8 enzyme complex, and the resulting products are named as precursor miRNAs (pre-miRNAs). Exportin-5, a nuclear transport protein, then mediates the exportation of pre-miRNAs to the cytoplasm, where they are sliced by nuclease Dicer yielding miRNA duplexes, or the mature miRNAs. The strand generally with the less-thermostable 5'-terminus incorporates into RNA-induced silencing complex (RISC), composed of the Argonaute (AGO) protein family and associated proteins, while the other strand is degraded. RISC with mature miRNA is able to recognize and bind to 3'UTR of a target mRNA and prompt mRNA cleavage with perfect complementarity or translational repression with imperfect complementarity.
The expression of target proteins reduces as a consequence of mRNA degradation or translational inhibition. MiRNAs thus are able to participate in diverse physiological processes, including tissue morphogenesis, cell proliferation, differentiation, apoptosis and different signaling pathways (8). Due to the low specificity of miRNA-mRNA interaction, each mRNA can be repressed by more than one miRNA (9), and a single miRNA can regulate multiple target mRNAs (10).
In contrast, miRNA dysregulation could result in various diseases. Neurological disorders, cardiovascular diseases, monogenic disorders, autoimmune diseases and cancer have been discovered to have a relationship with miRNA disruption (11).
Role of miRNAs in carcinogenesis and as cancer biomarkers
Both genetic and epigenetic changes through miRNA disruption engage in carcinogenesis. Genetically, miRNAs could downregulate tumor suppressive gene expression and upregulate oncogene expression (12). The same miRNA is able to have oncogenic activity in one particular cell type but a tumor-suppressive property in another based on different targets and pathogenesis (13). Under normal circumstances, miRNAs modulate the level of epigenetic regulatory proteins, such as DNA methyl transferases, histone deacetylases and components of the polycomb repressor complexes; they directly change the epigenetic status of promoter regions as well (14). Abnormal alterations of these epigenetic signatures by miRNA dysregulation are implicated in carcinogenesis.
However, first, the dysregulation of miRNAs concerns the major steps of the miRNA biogenesis as follows (15): genomic and genetic changes, such as deletions, mutations, amplifications, translocations and single nucleotide polymorphisms (SNPs); epigenetic alterations such as DNA methylation and histone acetylation; altered activity of transcription factor; and impaired expression or function of related proteins (Drosha, Dicer, Exportin-5, etc.).
Tumor tissues could be distinguished from normal tissues judging by the expression level of miRNAs. Differential expression of miRNAs is broadly revealed in lung cancer (16), colorectal cancer (17), renal cell carcinoma (18), prostate cancer (19), pancreatic cancer (20), ovarian cancer (21), etc. Moreover, specific miRNAs correspond to certain types of cancer and identify cancer tissue origins (22).
Obtaining tissue samples is much more inferior to non-invasive sampling, in terms of feasibility and safety, to detect the altered expression of miRNAs. It is observed that miRNAs are present in multiple human body fluids, such as plasma, urine, pleural fluid, colostrum, and cerebrospinal fluid (23). Additionally, it is well recognized that circulating miRNAs are correlated to tumors and are of diagnostic, prognostic, and monitoring values (24,25). Circulating miRNAs possess several advantages to distinguish themselves among cancer biomarkers. They are extraordinarily stable in body fluids, without the attack from endogenous RNases by the protection of vesicles or associated proteins (26). Both prolonged incubation at room temperature and multiple cycles of freeze-thawing exert minimal effects on miRNA amount (26). In addition, easy measurement provides sufficient dynamic data of the disease progression, response to treatments and cancer relapse, which help provide rational decision making for personalized therapies. Furthermore, circulating miRNAs overcome the problem of tumor heterogeneity because different parts of the tumor and metastatic sites all could release these pathologic signals into the circulation (27).
However, how do miRNAs gain access to the circulation? Basically, they enter via passive leakage and active secretion (24). The passive pathway, discharged from damaged and dead cells, however, plays a minor role in the production of circulating miRNAs. The active transport is mediated by microvesicles (microparticles and exosomes) and RNA-binding proteins, that is, AGO2 (28), high-density lipoproteins (29) and nucleophosmin (30). Tumor cells more preferentially excrete larger amount of exosomes than normal cells do, which presents the dysregulated status of tumor miRNAs in the circulation (31). The research of Taylor et al. revealed the levels of eight overexpressed miRNAs were similar between cellular and exosomal miRNAs and suggested circulating tumor exosomes as diagnostic biomarkers (32). This hypothesis is supported by some studies (33-35) but repudiated by other results (28,30). In spite of the contradictions of current findings, the use of circulating biomarkers as non-invasive cancer biomarkers is well established.
miRNAs as biomarkers in NSCLC and the mechanism
To date, studies have strongly demonstrated the role of miRNAs as biomarkers in NSCLC. Overexpression of oncogenic miRNAs and decreased expression of tumor suppressive miRNAs could both be detected in NSCLC. Some of them have been confirmed to be involved in the development or progression of lung cancer, and the principal miRNAs are miR-21, miR-17-92 cluster and miR-221/222 as oncogenic miRNAs and let-7 family, miR-34 family and miR-200 family as tumor suppressive miRNAs (36).
The let-7 family was the first discovered human-encoded miRNA, of which the expression was also shown reduced in NSCLC patients indicating poor prognosis (37,38). Let-7 possesses tumor suppressive activity, inhibiting multiple oncogenes such as RAS (39), MYC (40) and HMGA2 (41), and reduces the expression of cyclins (42). In lung cancer, chromosomal regions containing various let-7 genes were reported often deleted (43). Moreover, a frequent SNP at the let-7 complementary site 6 was validated to have an association with an increased risk for NSCLC among smokers (44).
The miR-34 family comprises miR-34a, miR-34b and miR-34c, acting as mediators of tumor suppression by P53 (45). All members of the miR-34 family are capable of repressing tumor growth and metastasis by targeting mRNAs participating in cell cycles, epithelial-mesenchymal transition (EMT), metastasis, stemness, apoptosis and senescence (46). It was observed that miR-34 genes were frequently downregulated by CpG methylation in various types of tumor or deleted as a minor cause (47). One study revealed that miR-34 synergistically with miR-15a/16 was significantly downregulated in NSCLC cell lines (48). Another study identified tissue miR-34a as an independent prognostic marker of recurrence in surgically resected NSCLC (49). Additionally, aberrant methylation of tissue miR-34 was indicated as a prognostic factor for NSCLC (50,51).
All five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) underwent remarkable downregulation in cells with EMT, which is regarded as a critical step in metastasis (52). EMT induced by the miRNAs was considered as a result of regulation of zinc finger E-box-binding homeobox (ZEB) transcription factors and E-cadherin (53). Loss of miR-200c expression was shown to give rise to an aggressive, invasive, and chemoresistant phenotype of NSCLC (54). However, other clinical outcomes contradict the above findings about miR-200c, as poor survival rates, not provided by previous studies, were demonstrated in NSCLC with overexpression of miR-200c (55,56). The oncogenic property of miR-200c was argued by its potential to target several tumor suppressor genes as a more dominant role than regulation of ZEB in NSCLC carcinogenesis (56).
MiR-21 is an oncogenic miRNA and overexpressed in multiple solid tumors (57), including NSCLC. MiR-21 promotes tumorigenesis through inhibition of regulators of the Ras/MEK/ERK pathway and blockage of apoptosis (58). Negative regulation on tumor suppressive genes, such as PTEN (59), MARCKS (60), PDCD4 (61) and TPM1 (62) has been reported to be part of miR-21’s oncogenic mechanism. The elevated expression of miR-21 was much higher in tumor tissues and cell lines with epidermal growth factor receptor (EGFR) mutation than those without the mutation, indicating that the EGFR signaling pathway increases the expression of miR-21 (63). To complicate the situation, downregulation of miR-21 inhibits the EGFR pathway, which implicates a feedback loop between miR-21 and EGFR (64,65). The research of Wei et al. identified plasma miR-21 as a sensitive and specific marker for early diagnosis for NSCLC and a predicative indicator for response sensitivity to platinum chemotherapy (66). Moreover, one retrospective analysis suggested the potential of miR-21 as a prognostic biomarker of early-stage lung adenocarcinoma (67).
More preferably in small cell lung cancer, the miR-17-92 cluster (miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a and miR-92a) was observed overexpressed with occasional gene amplification (68). The upregulated miR-17-92 cluster negatively regulates E2F (69), Myc (69), HIF-1α (70) and PTEN (71) as possible routes to apply its oncogenic function.
MiR-221 and miR-222 were shown particularly increased in tissues and cell lines of invasive and TNF-related apoptosis-inducing ligand (TRAIL) resistant NSCLC (72), through PTEN and TIMP3 downregulation (73). On the other hand, a relatively new study identified the growth suppressive ability of miR-221 and miR-222 in four other cell lines, which was different from the previous reports. This interesting finding might explore the dual roles of miRNAs not only in multiple cancer types but also in various cancer cell lines within one tumor class (74).
Clinical significance of miRNA biomarkers in NSCLC
One application of miRNAs is diagnosis of NSCLC, early diagnosis in particular. Additionally, the diagnostic use will be discussed separately in the latter part of the passage.
Interestingly, miRNAs could potentially differentiate lung cancer subtypes. For example, one study found four overexpressed miRNAs (miR-205, miR-93, miR-221 and miR-30e) in squamous cell carcinoma tissue and five more highly expressed miRNAs (miR-29b, miR-29c, let-7e, miR-100 and miR-125a-5p) in adenocarcinoma tissue (75). It may help with perioperative differentiation of histologic subtypes in those NSCLCs hard to perform a biopsy.
Additionally, miRNAs have been recognized of prognostic value in NSCLC. Surgical resection remains the only curative treatment, and survival is threatened by disease recurrence, which could be reflected by the level of miRNAs. Sanfiorenzo et al. identified two plasma miRNA panels associated with poor disease-free survival in resectable NSCLC, one of adenocarcinoma (high miR-155-5p, high miR-223-3p, and low miR-126-3p) and the other of squamous cell carcinoma (high miR-20a-5p, low miR-152-3p, and low miR-199a-5p) (76). According to a 24-microRNA panel from plasma, Sestini et al. classified 84 subjects into three groups of low, intermediate and high risk, which was subsequently found to be inversely correlated with 5-year survival (88.9%, 79.5% and 40.1%, respectively) (4). Among patients who underwent surgery, 76% of the 25 high-intermediate patients showed decreased risk calculated by the miRNA panel, while the risk first lowered but rebounded afterwards in one relapsing subject.
Furthermore, miRNAs could predict response to non-surgical therapies and the survival in NSCLC. Chen et al. enrolled 54 eligible patients of stage III or IV disease for radiotherapy and discovered four plasma miRNAs (hsa-miR-98-5p, hsa-miR-302e, hsa-miR-495-3p, and hsa-miR-613) with higher expression in responders (complete or partial response) than in non-responders (stable or progressive disease) (77). Additionally, the high miRNA level favored an objective response rate based on each cut-point. The first-line chemotherapy using cisplatin and vinorelbine was evaluated by a prospective study of biopsy tissue samples from 38 subjects (78). MiR-149 and miR-375 were discovered predictive for response and were also related to progression-free survival. Meanwhile, a prognostic score containing four miRNAs (miR-200c, miR-424, miR-29c and miR-124) was constructed and proposed to assess the prognosis in differentiating median survival times.
Except for the traditional treatments, miRNA biomarkers become involved in novel treatments as well, such as target treatment and immune therapy. One research study explored the predictive role of plasma miRNAs in first-line EGFR tyrosine kinase inhibitors (EGFR-TKIs) for advanced NSCLC patients with EGFR 19 deletion (79). Three miRNAs (miR-21, miR-27a, and miR-218) were verified with significantly high expression in the resistant group in contrast to the sensitive group. Another study found that high expression of miR-200c in tumor tissue exhibited a significant association with a higher disease control rate, longer progression-free survival and longer overall survival in advanced NSCLC patients with wild-type EGFR who received second- or third-line EGFR-TKIs, but no significant relation was found to patients with EGFR mutation (80). EMT induced by a low level of miR-200c was assumed to be the mechanism of less effective EGFR-TKIs in the wild-type EGFR population. Chen et al. showed that the expression of the miRNA-200 family in tumor tissue was associated with a level of programmed death-ligand 1 (PD-L1) (81). The regulation of PD-L1 through the miRNA-200/ZEB1 axis probably acts by targeting PD-L1 directly or suppressing EMT when high expression PD-L1 was demonstrated in the mesenchymal phenotype of NSCLC. Although lacking research evidence, miR-200 expression should serve as a promising predictor of anti-PD-1/PD-L1 therapy in NSCLC. Additional miRNAs of potentially predictive value for immune therapy include miR-34 (82) and miR-197 (83).
Media for measuring miRNAs
Sputum is rather a less studied area concerning miRNA biomarkers than plasma/serum. Sputum samples have benefits in that they contain the respiratory epithelial cells where the prolonged molecule genetic changes can be collected and tested (84). Nevertheless, there was doubt over the stability of miRNAs for high concentration of RNase activity in the sputum. However, it was proved that the miRNAs could be detectable up to 7 days after sputum collection with resistance of endogenous miRNAs to RNase activity (85). Another issue is that not all patients are able to expectorate sputum spontaneously, particularly former smokers and cancer-free patients. To address it, Su et al. proposed the application of lung fluke for sputum collection among those people without spontaneously expectorated sputum (86). Sputum samples would still be problematic for containing inconsistent cells in various samples, making miRNA quantification less convincing.
Whole blood miRNA examination was occasionally reported of diagnostic use in NSCLC, but a new study implied that whole blood miRNA expression might lack diagnostic value for NSCLC because the miRNA candidates did not differ in case and control groups or in pre-surgical and post-surgical groups (87). They explained the results by the fact that miRNAs exist extracellularly and within blood cells, and miRNA changes in whole blood may reflect other pathologic conditions and various physiologic statuses.
The role of serum and plasma as biomarkers of NSCLC, however, has been widely investigated. Additionally, serum and plasma are both accepted as better alternatives to sputum and whole blood, with greater specificity and sensitivity (88). Differences between serum and plasma were still observed, and the study of Wang et al. recommended plasma as the choice for studying circulating miRNAs since RNA released out of the cells during the coagulation process could conceal the true miRNA information (89).
miRNAs as biomarkers for early detection of NSCLC
Instead of a single miRNA, miRNA panels are more usually adopted to differentiate lung cancer from healthy controls in current studies, yielding higher sensitivity and specificity (90). However, no consensus of diagnostic panels has been achieved, and several meta-analyses tried to provide a general understanding about the diagnostic potential of miRNAs in NSCLC. Wang et al. conducted a meta-analysis of non-invasive miRNAs with a total of 28 articles, involving 2,121 NSCLC patients and 1,582 healthy controls (91). Multiple miRNA assays showed a pooled sensitivity of 0.83 and specificity of 0.82, which was more accurate than single miRNA assays with a sensitivity of 0.77 and specificity of 0.71. Another meta-analysis of non-invasive miRNAs performed by Chen et al. (20 articles, involving 1,563 NSCLC patients and 1,060 healthy controls) shared similar results (92)—the performance of multiple miRNAs (sensitivity and specificity of 0.81 and 0.84, respectively) was better than that of single miRNA assays (sensitivity and specificity of 0.73 and 0.77, respectively). Although the diagnostic value is displayed by the above meta-analyses, the overall sensitivity and specificity, both approximately 0.80, do not indicate excellent performance. Fortunately, there are individual studies showing promising results, as the details described in Table 1 depict. These studies all presented different miRNA panels or diagnostic models consisting of miRNAs and radiological features that could better detect early NSCLC, with some of them showing a lower false positive rate.
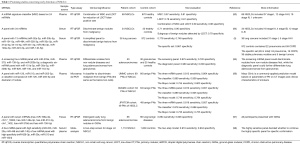
Full table
In 2011, Boeri et al. reported four ratio signatures of 24 miRNAs in plasma, regarding the risk to develop lung cancer, lung cancer diagnosis, risk of aggressive lung cancer and presence of aggressive lung cancer (99). The signatures were first identified by testing the tissues in the INT-IEO cohort as a training set and subsequently validated in the MILD cohort as validation set, while the two sets were from two LDCT screening trials. In 2014, based on the previously found 24 miRNAs, the same group constructed a miRNA signature classifier (MSC) of predictive, diagnostic and prognostic value (93). From 939 participants in the MILD trial, 69 of them with lung cancer and the remaining 870 as healthy controls, an analysis was performed. Interestingly, the combination of MSC and LDCT (double-positive result) gave rise to a five-fold reduction of LDCT false-positive rate from 19.4% to 3.7%, but unfortunately there was a decrease in sensitivity to 0.69. A non-randomized single-group trial, BIOMILD, is ongoing by this group, with the estimated completion date in January 2021; it concerns plasma miRNAs as a first-line screening test for lung cancer detection (http://ClinicalTrials.gov/, Identifier: NCT02247453). The intervention of plasma assay and other radiologic tests depend on the risk of the baseline miRNA profile, and the expected primary outcome is reduction of false positive cases in lung cancer detection through plasma miRNA profiling, with overall survival as a secondary outcome.
Bianchi et al. developed a panel of 34 miRNAs from serum for detection of early-stage NSCLC among asymptomatic high-risk individuals (94). Sera from the COSMOS trial were collected and divided into a training set and a testing group. The general sensitivity and specificity was 0.71 and 0.90 in the testing set, respectively. An extra analysis was performed addressing whether the predictor discriminated benign nodules detected by LDCT without cancer development from malignancy, and the specificity was 0.79 (26 out of 30 samples). By comparing a group of sera that were sampled before and after cancer onset, a significant elevation of average risk index was detected, indicating its function as a predictor of the conversion from a normal to a cancerous state. Four years later, the same group published a modified signature to 13 miRNAs and validated the new miRNA profiling in a large population of the COMOS trial (5). The sensitivity and specificity were 0.778 and 0.748, respectively, in a validation set of 1,008 subjects. Additionally, a specific set that included 83 participants with chronic obstructive pulmonary disease (COPD), stable pulmonary nodules, benign tumors or pneumonia displayed a rate of 0.867 (72 out of 83) with miRNA negative results. The overlap of five miRNAs between this 13-miRNA panel with the previous 24-miRNA panel encourages further study of the synergism towards increasing diagnostic accuracy.
Cazzoli et al. selected two panels of miRNAs from a sum of 742 miRNAs to screen and diagnose lung adenocarcinomas (95). They first tested a set of 30 plasma samples and later reevaluated a larger group of 105 samples, both composed of lung adenocarcinomas, lung granulomas, and healthy smokers. The proposed screening miRNA signature consisted of four microRNAs—miR-378a, miR-379, miR-139-5p, and miR-200b-5p—and could discriminate nodules from non-nodule diseases, with 0.975 sensitivity and 0.720 specificity. The six-miRNA diagnostic test (miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100, and miR-154-3p) divided the nodule diseases into lung adenocarcinomas and granulomas and showed 0.960 sensitivity and 0.600 specificity. One concern of this study is that the nodule group was solely composed of adenocarcinomas and granulomas and ruled out other benign nodules arbitrarily, which makes the result less reliable for advanced studies and clinical practices.
Lately, an integrated classifier combined by two miRNAs and one radiological feature was introduced for diagnosis of indeterminate primary nodules (PNs) (96). From 11 identified plasma miRNAs with significantly different expression, a panel of three miRNA biomarkers (miR-126, miR-210, and miR-205-5p) was used to distinguish malignant from benign PNs in a UMMC cohort (69 PNs of NSCLC and 66 benign PNs) at the beginning, generating a sensitivity of 0.812 and a specificity of 0.864. Additionally, univariate analysis determined that a history of cancer and smoking pack-years of the patients, and the diameter, spiculation, and upper lobe location of the nodules were associated with malignancy among clinical and radiological variables. Furthermore, the researchers grouped miR-126, miR-205-5p and the diameter of the nodule for a classifier design, which yielded 0.899 sensitivity and 0.909 specificity in the UMMC cohort. To completely compare the diagnostic value of the classifier, the Mayo Clinic model, a commonly applied prediction model based on parameters of PN on CT images and clinical characteristics of smokers, was tested in the same cohort, showing 0.754 sensitivity and 0.803 specificity. The classifier was validated in two extra cohorts, the BVAMC (63 PNs of NSCLC and 63 benign PNs) and JPHTCM cohorts (49 PNs of NSCLC and 49 benign PNs), generating similar results (BVAMC: 0.889 sensitivity and 0.905 specificity; JPHTCM: 0.878 sensitivity and 0.898 specificity). The three sets together confirmed a more accurate sensitivity and specificity of the classifier than those of the three-miRNA panel or the Mayo Clinic model (all P<0.05). This study was absolutely inspiring, and with high diagnostic values of the proposed classifier, it urges that supplementary large-scale validation tests and related clinical trials should be performed.
Increasing cases of ground-glass nodules (GGNs) appearing on CT images confuse doctors and intensify patients’ concern regarding the uncertainty of the nodules. A seven-miRNA panel was identified by He et al. with the aim of distinguishing early lung adenocarcinoma from benign nodules (97). The group of He et al. initially evaluated the miRNA expression of adenocarcinoma and adjacent non-para-tumor tissue in three subjects using next-generation sequencing (NGS) and the miRNAs of interest were validated in 73 lung adenocarcinomas by qRT-PCR. Out of the 23 altered miRNAs detected by NGS, seven miRNAs were confirmed to have significantly different expression (P<0.05). The panel was subsequently validated in 66 lung adenocarcinomas paired with 66 lung benign diseases, and it yielded a sensitivity of 0.864 and specificity of 0.606. One noteworthy consideration is that the miRNAs were measured from lung tissue, which is not an ideal approach for a non-invasive test, and further studies about circulating miRNAs are urgently needed. As a newly established field to differentiate GGNs, the miRNA panel has great potential, and the diagnostic specificity has room for improvement.
A very recent systematic review tried to resolve the inconsistency of proposed miRNA signatures for screening of stage I–II NSCLC (98). The included 20 studies encompassed 1,110 NSCLC patients and 1,009 controls in total. Four circulating miRNAs that were hardly influenced by hemolysis, each with high sensitivity (>0.80) and area under the curve (AUC) (>0.80), were identified as biomarkers of stage I–II NSCLC: miR-223, miR-20a, miR-448 and miR-145. Four more miRNAs—miR-628-3p, miR-29c, miR-210 and miR-1244—were proved to have high specificity (>0.90). Finally, a model of a two-step screening for stage I–II NSCLC was proposed, using the first four-miRNA panel as a sensitive test to decide whether to continue the second four-miRNA panel for a specific confirmation. This combined model, showing an estimate of more ideal sensitivity (0.916) and specificity (0.934), was assumed to be a candidate a preliminary screening test to LDCT for a cost-benefit consideration and minimal radiation exposure. For further validation of this screening model, an interventional research study with a large set is required.
Conclusions and perspectives
Currently, miRNAs have been increasingly recognized for their crucial role in tumorigenesis and have been extensively studied in the field of NSCLC, where their value as biomarkers is fully appreciated, in particular, their significance in diagnosis, prognosis and prediction of response to various treatments as described in this review. Among a great number of studies regarding the early detection of NSCLC by miRNAs, we discussed the remarkably meaningful and promising ones. Different miRNA panels and diagnostic models were proposed, with various diagnostic values reported, showing a bright future for a combined screening method for early NSCLC where the false positive rate is reduced.
However, problems yet remain. The inconsistency of miRNA panels prevents further comparison of diverse studies and decisions regarding an ideal diagnostic model. Additionally, the population of each study differs approximately, for example, in the various histological classifications of NSCLC, making the synthesis even unlikely. However, from another perspective, studies enrolling single types of NSCLC may gain more consistent results and could be conducted, supported by the literature that miRNA expression profiles could determine the histology of NSCLC (100,101). Some studies have already been performed, such as the two that only involved adenocarcinomas (95,97), but extra efforts are worth devoting to comprehensive and detailed investigations. Additionally, the combination of LDCT and miRNA assay needs more exploration for the purpose of ascertaining the imaging features that would be involved and how these two examinations could be balanced and coordinated to raise the diagnostic power. To be put into clinical practice, miRNAs still have a long way to go even if the inconsistency is solved. Once a promising model is established, large-scale clinical trials are required for validation and provision of a high level of evidence, which is the exact urgency of miRNA studies on biomarkers in NSCLC.
Acknowledgements
We would like to acknowledge professor Xianghuo He, from Fudan University Shanghai Cancer Center and Institutes of Biomedical Sciences, for his suggestions of revision.
Funding: This study was supported by the grant from Science and Technology Commission of Shanghai Municipality Medical Guidance Science&Technology Support Project (grant No. 16411966100), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (grant No. 20172005) and Shanghai Municipal Commission of Health and Family Planning Outstanding Academic Leaders Training Program (No. 2017BR055).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2014, National Cancer Institute. Bethesda, MD, based on November 2016 SEER data submission, posted to the SEER web site, April 2017. 2017. Available online: https://seer.cancer.gov/csr/1975_2014/
- National Lung Screening Trial Research T, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Sestini S, Boeri M, Marchiano A, et al. Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget 2015;6:32868-77. [Crossref] [PubMed]
- Montani F, Marzi MJ, Dezi F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Wang WT, Chen YQ. Circulating miRNAs in cancer: from detection to therapy. J Hematol Oncol 2014;7:86. [Crossref] [PubMed]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11:441-50. [Crossref] [PubMed]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev 2004;18:504-11. [Crossref] [PubMed]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008;9:102-14. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012;4:143-59. [Crossref] [PubMed]
- Giza DE, Vasilescu C, Calin GA. Key principles of miRNA involvement in human diseases. Discoveries (Craiova) 2014;2:e34. [Crossref] [PubMed]
- Malumbres M. miRNAs and cancer: an epigenetics view. Mol Aspects Med 2013;34:863-74. [Crossref] [PubMed]
- Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J 2012;18:215-22. [Crossref] [PubMed]
- Guan P, Yin Z, Li X, et al. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res 2012;31:54. [Crossref] [PubMed]
- Hamfjord J, Stangeland AM, Hughes T, et al. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One 2012;7:e34150. [Crossref] [PubMed]
- Chow TF, Youssef YM, Lianidou E, et al. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clin Biochem 2010;43:150-8. [Crossref] [PubMed]
- Carlsson J, Davidsson S, Helenius G, et al. A miRNA expression signature that separates between normal and malignant prostate tissues. Cancer Cell Int 2011;11:14. [Crossref] [PubMed]
- Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas 2013;42:67-71. [Crossref] [PubMed]
- Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007;67:8699-707. [Crossref] [PubMed]
- Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 2008;26:462-9. [Crossref] [PubMed]
- Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-41. [Crossref] [PubMed]
- Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 2012;32:326-48. [Crossref] [PubMed]
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087-92. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Esposito A, Criscitiello C, Locatelli M, et al. Liquid biopsies for solid tumors: Understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol Ther 2016;157:120-4. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 2010;38:7248-59. [Crossref] [PubMed]
- Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci 2013;14:14240-69. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Cheng L, Sharples RA, Scicluna BJ, et al. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles 2014;3. [Crossref] [PubMed]
- Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013;119:1159-67. [Crossref] [PubMed]
- Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679. [Crossref] [PubMed]
- Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med 2016;5. [Crossref] [PubMed]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [Crossref] [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. [Crossref] [PubMed]
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [Crossref] [PubMed]
- Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007;67:9762-70. [Crossref] [PubMed]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007;21:1025-30. [Crossref] [PubMed]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007;67:7713-22. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [Crossref] [PubMed]
- Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res 2008;68:8535-40. [Crossref] [PubMed]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 2010;17:193-9. [Crossref] [PubMed]
- Rokavec M, Li H, Jiang L, et al. The p53/miR-34 axis in development and disease. J Mol Cell Biol 2014;6:214-30. [Crossref] [PubMed]
- Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 2012;12:613-26. [Crossref] [PubMed]
- Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer 2011;10:55. [Crossref] [PubMed]
- Gallardo E, Navarro A, Vinolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009;30:1903-9. [Crossref] [PubMed]
- Kim YH, Lee WK, Lee EB, et al. Combined Effect of Metastasis-Related MicroRNA, miR-34 and miR-124 Family, Methylation on Prognosis of Non-Small-Cell Lung Cancer. Clin Lung Cancer 2017;18:e13-20. [Crossref] [PubMed]
- Wang Z, Chen Z, Gao Y, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther 2011;11:490-6. [Crossref] [PubMed]
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593-601. [Crossref] [PubMed]
- Hurteau GJ, Carlson JA, Roos E, et al. Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle 2009;8:2064-9. [Crossref] [PubMed]
- Ceppi P, Mudduluru G, Kumarswamy R, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res 2010;8:1207-16. [Crossref] [PubMed]
- Tejero R, Navarro A, Campayo M, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One 2014;9:e101899. [Crossref] [PubMed]
- Kim MK, Jung SB, Kim JS, et al. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch 2014;465:463-71. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010;18:282-93. [Crossref] [PubMed]
- Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 2010;411:846-52. [Crossref] [PubMed]
- Li T, Li D, Sha J, et al. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun 2009;383:280-5. [Crossref] [PubMed]
- Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27:2128-36. [Crossref] [PubMed]
- Zhu S, Si ML, Wu H, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 2007;282:14328-36. [Crossref] [PubMed]
- Seike M, Goto A, Okano T, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A 2009;106:12085-90. [Crossref] [PubMed]
- Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest 2010;90:144-55. [Crossref] [PubMed]
- Zhang KL, Han L, Chen LY, et al. Blockage of a miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in glioblastomas. Cancer Lett 2014;342:139-49. [Crossref] [PubMed]
- Wei J, Gao W, Zhu CJ, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer 2011;30:407-14. [Crossref] [PubMed]
- Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res 2011;17:1875-82. [Crossref] [PubMed]
- Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628-32. [Crossref] [PubMed]
- Aguda BD, Kim Y, Piper-Hunter MG, et al. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci U S A 2008;105:19678-83. [Crossref] [PubMed]
- Taguchi A, Yanagisawa K, Tanaka M, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res 2008;68:5540-5. [Crossref] [PubMed]
- Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology 2010;11:501-6. [Crossref] [PubMed]
- Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 2008;27:3845-55. [Crossref] [PubMed]
- Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009;16:498-509. [Crossref] [PubMed]
- Yamashita R, Sato M, Kakumu T, et al. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med 2015;4:551-64. [Crossref] [PubMed]
- Zhang YK, Zhu WY, He JY, et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol 2012;138:1641-50. [Crossref] [PubMed]
- Sanfiorenzo C, Ilie MI, Belaid A, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One 2013;8:e54596. [Crossref] [PubMed]
- Chen X, Xu Y, Liao X, et al. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol 2016;37:11927-36. [Crossref] [PubMed]
- Berghmans T, Ameye L, Willems L, et al. Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer 2013;82:340-5. [Crossref] [PubMed]
- Wang S, Su X, Bai H, et al. Identification of plasma microRNA profiles for primary resistance to EGFR-TKIs in advanced non-small cell lung cancer (NSCLC) patients with EGFR activating mutation. J Hematol Oncol 2015;8:127. [Crossref] [PubMed]
- Li J, Li X, Ren S, et al. miR-200c overexpression is associated with better efficacy of EGFR-TKIs in non-small cell lung cancer patients with EGFR wild-type. Oncotarget 2014;5:7902-16. [Crossref] [PubMed]
- Chen L, Gibbons DL, Goswami S, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 2014;5:5241. [Crossref] [PubMed]
- Cortez MA, Ivan C, Valdecanas D, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2015;108. [Crossref] [PubMed]
- Fujita Y, Yagishita S, Hagiwara K, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther 2015;23:717-27. [Crossref] [PubMed]
- Thunnissen FB. Sputum examination for early detection of lung cancer. J Clin Pathol 2003;56:805-10. [Crossref] [PubMed]
- Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 2010;67:170-6. [Crossref] [PubMed]
- Su J, Anjuman N, Guarnera MA, et al. Analysis of Lung Flute-collected Sputum for Lung Cancer Diagnosis. Biomark Insights 2015;10:55-61. [Crossref] [PubMed]
- Patnaik SK, Kannisto ED, Mallick R, et al. Whole blood microRNA expression may not be useful for screening non-small cell lung cancer. PLoS One 2017;12:e0181926. [Crossref] [PubMed]
- Gyoba J, Shan S, Roa W, et al. Diagnosing Lung Cancers through Examination of Micro-RNA Biomarkers in Blood, Plasma, Serum and Sputum: A Review and Summary of Current Literature. Int J Mol Sci 2016;17:494. [Crossref] [PubMed]
- Wang K, Yuan Y, Cho JH, et al. Comparing the MicroRNA spectrum between serum and plasma. PLoS One 2012;7:e41561. [Crossref] [PubMed]
- Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts 2017;8:61-81. [Crossref] [PubMed]
- Wang H, Wu S, Zhao L, et al. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology 2015;20:56-65. [Crossref] [PubMed]
- Chen L, Jin H. MicroRNAs as novel biomarkers in the diagnosis of non-small cell lung cancer: a meta-analysis based on 20 studies. Tumour Biol 2014;35:9119-29. [Crossref] [PubMed]
- Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 2014;32:768-73. [Crossref] [PubMed]
- Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503. [Crossref] [PubMed]
- Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. [Crossref] [PubMed]
- Lin Y, Leng Q, Jiang Z, et al. A classifier integrating plasma biomarkers and radiological characteristics for distinguishing malignant from benign pulmonary nodules. Int J Cancer 2017;141:1240-8. [Crossref] [PubMed]
- He Y, Yang Y, Kuang P, et al. Seven-microRNA panel for lung adenocarcinoma early diagnosis in patients presenting with ground-glass nodules. Onco Targets Ther 2017;10:5915-26. [Crossref] [PubMed]
- Moretti F, D'Antona P, Finardi E, et al. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget 2017;8:94980-96. [PubMed]
- Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713-8. [Crossref] [PubMed]
- Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430-41. [Crossref] [PubMed]
- Bishop JA, Benjamin H, Cholakh H, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res 2010;16:610-9. [Crossref] [PubMed]


