Altered expression of TIAM1 in endotoxin-challenged airway epithelial cells and rodent septic models
Introduction
Sepsis is a leading cause of death in critically ill patients. However, despite modern advances, mortality rate remains high (29%) (1) with increasing incidence which was about 13% annually (2) during the past decade in US, thus demanding ongoing attention and research. Better understanding of the underlying pathogenesis could help clinician to recognize the manifestation of sepsis, along with the early management strategies, to expeditiously provide appropriate care and decrease resultant morbidity and mortality.
The pathogenesis of sepsis is complex and multifactorial, exaggerated inflammatory processes, inappropriate regulation of cellular reactions and coagulation abnormalities are involved (3). In current opinion, sepsis is thought to be a series of complicated immune responses triggered by pathogen, which are characterized by continued activation of neutrophils and macrophages/monocytes, upregulation of lymphocyte costimulatory molecules, rapid lymphocyte apoptosis and enhanced necrosis of cells/tissues. The resulting imbalance of these immune mechanisms may lead to either excess tissue damage or immunosuppression (4). Several signal transduction pathways are involved in these processes and reactions. Signal transduction through Toll-like receptors (TLRs) leads to the activation of transcription factors, such as nuclear factor-kappa B (NF-κB) and activator protein 1 (AP-1), and the expression of genes related to the inflammatory response (5,6). The mitogen-activated protein kinase (MAPK) pathways are also activated, resulting in the release of numerous inflammatory mediators (7,8). These pathways are closely tied to the inflammatory response, with crosstalk driving the deregulated response.
In our previous study (9) utilizing bioinformatics approaches, we have identified the differential expression of genes from multiple microarray studies of whole blood samples of human sepsis (10-15). After clustering analysis using Kyoto Encyclopedia of Genes and Genomes (KEGG), we found these genes are enriched in “Regulation of actin cytoskeleton” pathway, which might play a previously under-recognized role in human sepsis. In sepsis, reorganization of the actin cytoskeleton in the epithelium during inflammation will lead to breakdown of epithelial barrier integrity (16), increasing epithelial permeability and pathogenic microorganisms invasion. Epithelial barrier dysfunction is thought as a key factor to explain the pathogenesis of multiple organ dysfunctions at the cellular level (17). These same derangements can also be found in endothelial cells, contributing to severity of microvascular leak, which manifests as tissue and organ edema, hypotension, and shock (18).
In this study, we focused on one of the components of “Regulation of actin cytoskeleton”, T-lymphoma invasion and metastasis 1 (TIAM1), a guanine nucleotide exchange factor (GEF) specific for Rac1, Cdc42 and RhoA activation (19). The expression levels of TIAM1 have been associated with mortality among sepsis patients (20). We further validated the expression of TIAM1 using both in vivo and in vitro models of sepsis. Our findings have the potential for the development of diagnostic and treatment strategies that are relevant for patient management.
Methods
Epithelial cell culture
Primary human tracheobronchial epithelial cells (HTBEs) were collected from cadaveric lung donors (no history of respiratory disease) and grown at the air-liquid interface (ALI) as described previously (21). Cells were used after 6 weeks in ALI culture and treated with 1 µg/mL lipopolysaccharide (LPS) (0111:B4; Sigma, St. Louis, MO). Cells were harvested at 3, 6, 24 and 48 h after treatment. The control groups received phosphate-buffered saline (PBS) and were harvested at the above time points.
The human bronchial epithelial cell line Beas-2B was purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM/F12 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) and Penicillin-Streptomycin (Invitrogen, Carlsbad, CA). Cells were used after reaching 80% confluence and treated with 1 µg/mL LPS. Cells were harvested at 3, 6 and 12 h after treatment.
LPS challenged mice
Male C57BL/6J (wild type, WT) mice aged 10–12 weeks (Jackson Laboratory, ME) were randomly exposed to intravenous PBS or LPS (0127:B8; Sigma, St. Louis, MO) 9.5 mg/kg via retro-orbital injection (22) for up to 24 hours (n=6 for each group). Lung tissue was collected at 4 or 24 h after injection and stored at −80 °C for mRNA and protein isolation.
Colon ascendens stent peritonitis (CASP) induced septic rats
Male SD rats (weight 200–300 g) were purchased from Sun Yat-sen University (Guangzhou, China). CASP was induced as described previously (23). Briefly, rats were intraperitoneally anesthetized by ketamine/xylazine (50 mg/kg). The ascending colon was brought out through a 2-cm midline incision. A 16G ×3 mm stent was placed 1.5 cm distal to the ileocecal valve and fixated to the wall with sutures. Gentle pressure was applied to express a 2-mm bead of colon content from the patent stent and the ascending colon was returned to the lower left abdominal quadrant. Normal saline (10 mL/kg) was administered subcutaneously after the closure of peritoneal cavity as fluid resuscitation. The Sham group underwent the same procedure (laparotomy and ascending colon exposure) without stent insertion (n=5 for each group). Lung tissue was collected at 3 or 6 h after CASP surgery and used for protein isolation. The whole blood samples were collected from inferior vena cava and used for mRNA isolation.
Measurements of HTBEs barrier function
We used a Countess™ Automated Cell Counter (Invitrogen, Carlsbad, CA) to quantify cell viability. Briefly, 10 µL 0.4% Trypan blue stain was added to 10 µL cells after harvest and mixed gently, then 10 µL of the mixture was added to the slide and counted by the machine. We calculated percentage of dead cells by the following formula: Dead cell percentage =(1 – viability) ×100%. To monitor LPS induced changes of ionic permeability on all monolayers of HTBEs, upon reaching 85–90% confluency the cells were grown at the ALI until maturity and confluence were reached (5–6 weeks), then we measured transepithelial electrical resistance (TER) with an epithelial voltohmmeter (World Precision Instruments, Sarasota, FL) as detailed in the manufacturer’s instructions. Cultures with TERs above 250 Ω·cm2 were used for exposures. In order to obtain a representative sample, TER was measured on three different areas of the monolayer. Additionally, macromolecular permeability was measured as previously described (24), 0.5 mL of 10 mM Alexafluor 647-BSA (bovine serum albumin) conjugate (Invitrogen, Carlsbad, CA) was added to the apical side of cultures, then basolateral medium was changed to 2 mL PBS, the transwell cultures were incubated for 60 min at 37 °C, fluorescent quantification in PBS was performed by fluorescent volume analysis (voxels) after normalization to stain controls lacking Alexafluor 647. Cells on transwell chambers were subsequently washed twice with PBS and fixed for 20 min at room temperature using 3% sucrose/3% paraformaldehyde in PBS. After fixing, cells were washed once with PBS and permeabilized for 15 min with 0.1% Triton X-100 in PBS and then stained with 0.3 mM 4,6-diamino-2-phenyl indole (DAPI) (Molecular Probes, Eugene, OR) in PBS on ice for 1h, Membranes were cut from the transwell frames with a scalpel, rinsed in milli-Q water, mounted cells-up in glycerol containing 100 mM Tris (pH 8.0) and stored at −20 °C until examined. Images of leaks were captured by fluorescence microscopy (OLYMPUS BX50WI, Tokyo, Japan; magnification was 20×). The averages of at least 3 independent experiments were shown.
PCR Array and qRT-PCR
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) for mice lung tissue, cDNA was prepared using high capacity cDNA archive kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s guide. Quantitative real-time PCR was performed with the TaqMan Genotyping Master Mix (Invitrogen, Carlsbad, CA) with the following conditions: (50 °C, 2 min; 95 °C, 10 min; 95 °C, 15 s; 60 °C, 60 s) ×40. Real-time quantitative RT-PCR results were normalized against an internal control (GAPDH). Using primers for mouse TIAM1 (Mm01170430_m1) and GAPDH (Mm9999915_g1) purchased from TaqMan. The single gene quantitative reverse transcription polymerase chain reaction (qRT-PCR) with TaqMan universal PCR master mix (Invitrogene, Carlsbad, CA, USA) was performed on an ABI Prism® 7300 Sequence Detection System, using a fluorogenic 5’ nuclease assay (Applied Biosystems, Foster City, CA, USA). Assays were performed in triplicate.
Total RNA of the whole blood of rats was extracted using RNApure blood kit (CWBIO, Beijing, China) following the manufactures’ instructions, cDNA was synthesized from total RNAs using 1st cDNA synthesis kit (TAKALA, JPN). qPCR amplifications were set up in a 96-well plate and run on CFX96 Touch™ Real-Time PCR detection system (Bio-rad, Hercules, CA, USA). The parameters were 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 30 s for 40 cycles. Amplification data were quantified using the comparative threshold-cycle method. As a loading control, β-actin cDNA was also amplified with primer. Using primers for rat TIAM1 (NM_001100558.1) purchased from GeneCopoeia, β-actinF: 5'-CACCCGCGAGTACAACCTTC-3', β-actinR: 5'-CCCATACCCACCATCACACC-3'. Assays were performed in triplicate.
Western blot analysis
We further validated protein expression of TIAM1 using Western blots in both cells and animal samples. Homogenized tissue or cells were lysed in lysis buffer (Cell Signaling, Danvers, MA, USA) plus 1 × Protease/Phosphatase Inhibitor Cocktail (Cell Signaling, Danvers, MA). The protein concentration was determined by BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Equal amounts of total protein (30 µg) were separated on 4–20% Tris-Glycine gels (Invitrogen, Carlsbad, CA) followed by blotting to nitrocellulose membrane (BioRad, Hercules, CA). Membranes were blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween 20 for 45 min and probed with primary antibodies: anti-TIAM1 IgM primary antibody (sc-6486, Santa Cruz, CA); anti-β-actin (4967S, Cell Signaling, Danvers, MA); anti-GAPDH (ab8245, Abcam, Cambridge, MA). Overnight at 4 °C followed by a goat anti-mouse IgM-HRP (horseradish peroxidase) secondary antibody (sc-2064, Santa Cruz, CA); a goat anti-rabbit IgG-HRP (horseradish peroxidase) secondary antibody (656120, Invitrogen, Carlsbad, CA); a goat anti-mouse IgG-HRP (horseradish peroxidase) secondary antibody (626520, Invitrogen, Carlsbad, CA), chemiluminescent detection (Thermo Fisher Scientific, Waltham, MA) was employed for band detection.
Statistical analysis
Our qRT-PCR data analysis was based on the 2–ΔΔCt method. Briefly, the 3–6 replicate Ct values for each gene and sample were averaged, and then the –ΔCt value for targeted genes in each sample was calculated using the average of the Ct values for 2 endogenous control genes (GAPDH or β-actin) (25). The expression level of target gene (relative to the average of these reference genes) was then calculated as 2–ΔΔCt. NIH ImageJ software (NIH, Bethesda, MD, USA) (26) was used for quantification of the density of bands on a western blot and β-actin or GAPDH was used for normalization. Data are presented as mean ± SEM of at least 3–6 experiments per condition. The SPSS 13.0 software (SPSS Inc., Chicago, IL, USA) was used for the data analysis. Intergroup differences were evaluated using one-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test for >2 group comparisons, or two-sided Student’s t-test for two groups. P<0.05 was considered statistically significant.
Results
LPS-induced barrier dysfunction in HTBEs
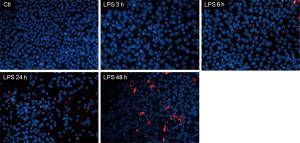
The percentage of dead cells was significantly increased after LPS stimulation at 3 h, 24 h (from 18.0%±2.1% to 35.0%±2.8% and 15.0%±3.5% to 37.5%±3.2% respectively, n=3 per group, P<0.05) (Figure 1A,B). TER, a measure of ionic permeability, was decreased at 3 and 48 h after LPS stimulation when compared to control group (from 276.2±14.8 to 206.7±14.5 Ω·cm2 and 371.0±22.9 to 289.6±17.5 Ω·cm2 respectively, n=3 per group, P<0.05) (Figure 2A). In addition, we measured fluorescently labeled BSA in basolateral PBS to represent macromolecular permeability of epithelial monolayers by comparing the LPS treatment group to the control, which was normalized to 100%. The level of fluorescently labeled BSA was increased at 3, 6, 24 and 48 h (n=3 per group, P<0.05) showing an increasing trend (Figure 2B). Moreover, fluorescent BSA leakage in the apical compartment occurred at 6 h and continually increased at late time points (Figure 3).
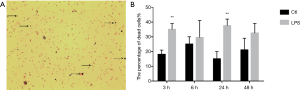

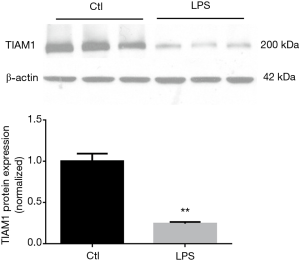
The protein level of TIAM1 was decreased after LPS stimulation in Bease-2B cells
We examined TIAM1 protein expression in Bease-2B cells and found that after LPS challenge, it was decreased at 3 h (n=3 per group, P<0.01) (Figure 4). The finding revealed that the protein level of TIAM1 was decreased after LPS stimulation in epithelium at early time point.
The expression of TIAM1 mRNA and protein was decreased in lungs of septic model
We further measured TIAM1 expression in rodent septic models. We observed down-regulation of TIAM1 mRNA in lungs of LPS challenged mice when compared to the controls at 4 and 24 h, fold change was −2.38 and −11.68, respectively (n=6 per group, P<0.05) (Figure 5A). The protein expression of TIAM1 was also significantly decreased at 24 h in lung tissue of LPS challenged mice (n=6 per group, P<0.01) (Figure 5B). Similar changes were found in lung tissue of CASP-induced septic rats, where the protein expression of TIAM1 was decreased at both 3 and 6 h (n=5 per group, P<0.05) (Figure 6A), however, the expression of TIAM1 had an increasing trend at 6 h time point when compared with which at 3 h, but there was no significant change between the two groups.
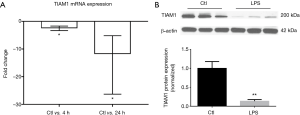
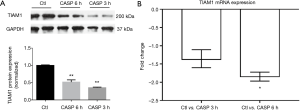
The blood mRNA level of TIAM1 was decreased after CASP in rats
We also examined TIAM1 gene expression in whole blood from septic rats. Interestingly, we found the mRNA expression of TIAM1 in peripheral blood was decreased after CASP in rats at 6 h (fold change =−1.85, n=5 per group, P<0.05) (Figure 6B).
Discussion
The actin-cytoskeleton is considered as the backbone of cells allowing mobility and migration of cells within the body. In carcinogenesis, the ability of cells to invade and migrate past the basement membrane may be dependent on conformational changes within this cellular backbone. The regulation of actin cytoskeleton pathway has already been demonstrated its importance in invasive metastatic sporadic colorectal cancer (27). In current opinion, breakdown in epithelial and endothelial barrier function plays a crucial role in the pathogenesis of sepsis (17,18). Regulation of actin cytoskeleton pathway participates in the whole process of barrier integrity, changes of cell junctions, rearrangement of the cell cytoskeleton and modulation of cell activation (16). Component such as myosin light chain kinase (MLCK) in regulation of actin cytoskeleton pathway is involved in LPS-induced disruption of epithelial permeability (28,29). LPS exposure could stimulate MLC phosphorylation, and induce actin rearrangement in human airway epithelial cell.
In our study we evaluated damaged barrier function induced by LPS in primary cultured HTBEs, our results suggested that LPS-induced barrier dysfunction in HTBEs is a valid in vitro model that mimics changes in the lung during human sepsis, and then we focused on examining the differential expression of TIAM1. As an upstream GEF of Rac1, TIAM1 is mainly involved in the regulation of Rac1 mediated signaling pathways including cytoskeletal activities, cell polarity, endocytosis and membrane trafficking, cell migration, adhesion and invasion, cell growth and survival, metastasis and carcinogenesis (30).
Recently, many studies have begun to investigate the role of TIAM1/Rac1 in the epithelial cell barrier. A study revealed that knock-down (KD) of TIAM1 by siRNA in epithelial cells led to the disassembly of cell-cell adhesions and TIAM1-Rac signaling is essential for the maintenance of E-cadherin mediated cell-cell adhesions (31). Junction maturation and tight junction (TJ) barrier function were severely impaired in TIAM1-deficient cells, re-expression of full-length TIAM1 or constitutively active V12Rac1 in TIAM1KD epithelial cells led to complete maturation of intercellular adhesions and rescued the impaired TJ formation (32). The same phenomenon was also found that depletion of paracingulin (CGNL1), a TIAM1-interacting protein, decreased accumulation of TIAM1 at epithelial junctions, down-regulates Rac1 activity and then affected TJ barrier development during TJ assembly by the calcium-switch (33), increased Rac1 activity driven by TIAM1 overexpression can rescue the TER phenotype of CGNL1(−) cells. These results all showed that normal levels of TIAM1 are required to ensure efficient junction assembly and may enhance the epithelial barrier function. All these studies were limited to in vitro epithelial cells, and lack of determination of the expression of TIAM1 during sepsis.
In our in vitro study, we detected altered protein expression of TIAM1, which was decreased after LPS stimulation in Bease-2B cells. In our in vivo study the mRNA expression of TIAM1 was significantly decreased by LPS intravenous injection when compared with the control group (the fold changes were −2.38 and −11.68 at 4 and 24 h, respectively). The protein expression of TIAM1 in the lung of septic rats was also decreased dramatically, but there was an increasing trend at 6 h time point compared to the levels at 3 h, we thought it represented kinetics of TIAM1 in response to septic insult which may reflect a protection mechanism, further investigation should be conducted to explain the exact response of this trend. Decreased expression of TIAM1 could affect the interaction of TIAM1/Rac1 signaling, and cytoskeletal rearrangement, causing junction disassembly, loss of polarity, epithelial barrier dysfunction and promote the development of sepsis. It suggests that the epithelial injury in sepsis is not only an explanatory pathophysiologic event, but also serves as indicators of disease and potential targets for future therapies. Several promising epithelial biomarkers have been identified in recent studies: Krebs von den Lungen-6 (KL-6), receptor for advanced glycation end products (RAGE), surfactant proteins, club cell protein 16 (CC16), and soluble Fas/FasL (34). These biomarkers can be used to assess lung epithelial integrity and injury. The results presented here also suggest the potential of utilizing TIAM1 to identify organ dysfunction earlier, predict outcome, and even to test novel therapeutic interventions.
There are limitations in our study. First, the underlying mechanism of TIAM1 downregulation in sepsis remains unclear. In carcinogenesis, phosphorylation of TIAM1 at Y384 is induced by Src activity and is associated with reduced total TIAM1 level then adherent junction (AJ) disassembly and cell migration (35). TIAM1 can also be ubiquitylated and degraded by the E3 ubiquitin ligase HUWE1 that facilitates junction disassembly, migration, and invasion (36). MEK-ERK signal pathway is also participated in TIAM1 degradation (37). The downregulation of TIAM1 in septic context need further investigation. Second, we just measured the total TIAM1 expression in epithelial cells, but the localization of TIAM1 and Rac1 along the apicobasal is different, thus generating an apicobasal Rac activity gradient at developing cell-cell junctions (38). In mechanically stretched epithelial cells TIAM1 is translocated from the membrane to the cytosolic fraction, this translocation of TIAM1 correlated with reduced Rac1 activity, decreased lamellipodial extensions, and inhibition of cell migration (39). Therefore, the exact localization of TIAM1 in epithelial cells in septic context warrants further study.
Most importantly, a human study aimed to study septic shock adults reported that TIAM1 gene expression was increased significantly in survivors compared to non-survivors by microarray findings (20), suggesting expression levels of TIAM1 could serve as a prognostic indicator for sepsis. Thus, given that peripheral blood can serve as an accessible surrogate tissue outside of the lung for noninvasive discovery of molecular biomarkers for sepsis, further investigation of its expression in peripheral blood of sepsis patients is warranted.
In summary, our study has gained novel insights on sepsis pathogenesis and confirmed significant changes of genes in the “Regulation of actin cytoskeleton” pathway, such as TIAM1. Furthermore, TIAM1 may serve as a prognostic indicator and novel target for effective antisepsis therapeutics.
Acknowledgements
Funding: This work was supported in part by NIH Grant no. 1R21AI110844 (to Li Gao) and Grant no. 81071536 from the National Natural Science Foundation of China (to X Guan).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The Johns Hopkins University Institutional Animal Care and Use Committee approved the protocols involving mice. The Sun Yat-sen University Animal Care and Use Committee approved the protocols involving rats.
References
- Stevenson EK, Rubenstein AR, Radin GT, et al. Two decades of mortality trends among patients with severe sepsis: a comparative meta-analysis*. Crit Care Med 2014;42:625-31. [Crossref] [PubMed]
- Gaieski DF, Edwards JM, Kallan MJ, et al. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med 2013;41:1167-74. [Crossref] [PubMed]
- Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med 2013;369:840-51. [Crossref] [PubMed]
- Stearns-Kurosawa DJ, Osuchowski MF, Valentine C, et al. The pathogenesis of sepsis. Annu Rev Pathol 2011;6:19-48. [Crossref] [PubMed]
- Salomao R, Martins PS, Brunialti MK, et al. TLR signaling pathway in patients with sepsis. Shock 2008;30 Suppl 1:73-7. [Crossref] [PubMed]
- Castoldi A, Braga TT, Correa-Costa M, et al. TLR2, TLR4 and the MYD88 signaling pathway are crucial for neutrophil migration in acute kidney injury induced by sepsis. PLoS One 2012;7. [Crossref] [PubMed]
- Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol 2002;20:55-72. [Crossref] [PubMed]
- Cornell TT, Rodenhouse P, Cai Q, et al. Mitogen-activated protein kinase phosphatase 2 regulates the inflammatory response in sepsis. Infect Immun 2010;78:2868-76. [Crossref] [PubMed]
- Ma J, Chen C, Barth AS, et al. Lysosome and Cytoskeleton Pathways Are Robustly Enriched in the Blood of Septic Patients: A Meta-Analysis of Transcriptomic Data. Mediators Inflamm 2015;2015. [Crossref] [PubMed]
- Shanley TP, Cvijanovich N, Lin R, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med 2007;13:495-508. [Crossref] [PubMed]
- Cvijanovich N, Shanley TP, Lin R, et al. Validating the genomic signature of pediatric septic shock. Physiol Genomics 2008;34:127-34. [Crossref] [PubMed]
- Pankla R, Buddhisa S, Berry M, et al. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol 2009;10:R127. [Crossref] [PubMed]
- Bermejo-Martin JF, Martin-Loeches I, Rello J, et al. Host adaptive immunity deficiency in severe pandemic influenza. Crit Care 2010;14:R167. [Crossref] [PubMed]
- Sutherland A, Thomas M, Brandon RA, et al. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit Care 2011;15:R149. [Crossref] [PubMed]
- Banchereau R, Jordan-Villegas A, Ardura M, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One 2012;7. [Crossref] [PubMed]
- Ivanov AI, Parkos CA, Nusrat A. Cytoskeletal regulation of epithelial barrier function during inflammation. Am J Pathol 2010;177:512-24. [Crossref] [PubMed]
- Fink MP, Delude RL. Epithelial barrier dysfunction: a unifying theme to explain the pathogenesis of multiple organ dysfunction at the cellular level. Crit Care Clin 2005;21:177-96. [Crossref] [PubMed]
- Goldenberg NM, Steinberg BE, Slutsky AS, et al. Broken barriers: a new take on sepsis pathogenesis. Sci Transl Med 2011;3. [Crossref] [PubMed]
- Habets GG, Scholtes EH, Zuydgeest D, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell 1994;77:537-49. [Crossref] [PubMed]
- Pachot A, Lepape A, Vey S, et al. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett 2006;106:63-71. [Crossref] [PubMed]
- Karp PH, Moninger TO, Weber SP, et al. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol 2002;188:115-37. [PubMed]
- Damarla M, Parniani AR, Johnston L, et al. Mitogen-activated protein kinase-activated protein kinase 2 mediates apoptosis during lung vascular permeability by regulating movement of cleaved caspase 3. Am J Respir Cell Mol Biol 2014;50:932-41. [Crossref] [PubMed]
- Lustig MK, Bac VH, Pavlovic D, et al. Colon ascendens stent peritonitis--a model of sepsis adopted to the rat: physiological, microcirculatory and laboratory changes. Shock 2007;28:59-64. [Crossref] [PubMed]
- Olivera DS, Boggs SE, Beenhouwer C, et al. Cellular mechanisms of mainstream cigarette smoke-induced lung epithelial tight junction permeability changes in vitro. Inhal Toxicol 2007;19:13-22. [Crossref] [PubMed]
- Cheadle C, Berger AE, Mathai SC, et al. Erythroid-specific transcriptional changes in PBMCs from pulmonary hypertension patients. PLoS One 2012;7. [Crossref] [PubMed]
- Budisavljevic M, Ronco PM, Verroust PJ, Angiotensin II. AII)-related idiotypic network. III. Comparative analysis of idiotopes and paratopes borne by monoclonal antibodies raised against AII (AB1) and its internal image (AB3). J Immunol 1990;145:1440-9. [PubMed]
- Kanaan Z, Qadan M, Eichenberger MR, et al. The actin-cytoskeleton pathway and its potential role in inflammatory bowel disease-associated human colorectal cancer. Genet Test Mol Biomarkers 2010;14:347-53. [Crossref] [PubMed]
- Eutamene H, Theodorou V, Schmidlin F, et al. LPS-induced lung inflammation is linked to increased epithelial permeability: role of MLCK. Eur Respir J 2005;25:789-96. [Crossref] [PubMed]
- Moriez R, Salvador-Cartier C, Theodorou V, et al. Myosin light chain kinase is involved in lipopolysaccharide-induced disruption of colonic epithelial barrier and bacterial translocation in rats. Am J Pathol 2005;167:1071-9. [Crossref] [PubMed]
- Boissier P, Huynh-Do U. The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling. Cell Signal 2014;26:483-91. [Crossref] [PubMed]
- Malliri A, van Es S, Huveneers S, et al. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J Biol Chem 2004;279:30092-8. [Crossref] [PubMed]
- Mertens AE, Rygiel TP, Olivo C, et al. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J Cell Biol 2005;170:1029-37. [Crossref] [PubMed]
- Guillemot L, Paschoud S, Jond L, et al. Paracingulin regulates the activity of Rac1 and RhoA GTPases by recruiting Tiam1 and GEF-H1 to epithelial junctions. Mol Biol Cell 2008;19:4442-53. [Crossref] [PubMed]
- Chawla LS, Fink M, Goldstein SL, et al. The Epithelium as a Target in Sepsis. Shock 2016;45:249-58. [Crossref] [PubMed]
- Woodcock SA, Rooney C, Liontos M, et al. SRC-induced disassembly of adherens junctions requires localized phosphorylation and degradation of the rac activator tiam1. Mol Cell 2009;33:639-53. [Crossref] [PubMed]
- Vaughan L, Tan CT, Chapman A, et al. HUWE1 ubiquitylates and degrades the RAC activator TIAM1 promoting cell-cell adhesion disassembly, migration, and invasion. Cell Rep 2015;10:88-102. [Crossref] [PubMed]
- Magliozzi R, Kim J, Low TY, et al. Degradation of Tiam1 by casein kinase 1 and the SCFbetaTrCP ubiquitin ligase controls the duration of mTOR-S6K signaling. J Biol Chem 2014;289:27400-9. [Crossref] [PubMed]
- Mack NA, Porter AP, Whalley HJ, et al. beta2-syntrophin and Par-3 promote an apicobasal Rac activity gradient at cell-cell junctions by differentially regulating Tiam1 activity. Nat Cell Biol 2012;14:1169-80. [Crossref] [PubMed]
- Desai LP, Chapman KE, Waters CM. Mechanical stretch decreases migration of alveolar epithelial cells through mechanisms involving Rac1 and Tiam1. Am J Physiol Lung Cell Mol Physiol 2008;295:L958-65. [Crossref] [PubMed]


