Functional miRNA variants affect lung cancer susceptibility and platinum-based chemotherapy response
Introduction
Lung cancer is the most frequently diagnosed cancer and the leading cause of cancer related death in the world (1). In China, over the past 30 years, lung cancer has taken the place of liver cancer as the highest fatality rate of malignant tumors, becoming the top of the list of cancer deaths. According to the World Health Organization (WHO) “Histological Typing of Lung and Pleural Tumours”, depending on the degree of differentiation and morphological characteristics, lung cancer can be classified into non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC). Over 80% are NSCLC patients, which comprise squamous-cell carcinoma (SCC), adenocarcinoma (ADC), adenosquamous carcinoma (AC), and large-cell carcinoma. SCLC is comparatively simple, but grows quickly and has a tendency of early transfer. Surgery is the mainstay of therapy for early stage of lung cancer, while most of lung cancer (over 70%) typically treated by platinum-based chemotherapy, are advanced or metastatic at the time of diagnosis (2). But large individual differences occurs in the chemotherapy efficacy of platinum-based drugs with the overall 5-year relative survival for all lung cancer patients remaining low (15%) and the recurrence rate still high, even in early-stage groups (3-6). Therefore, there is urgent need for the early prediction and therapeutic strategy guideline for lung cancer, which underlines the importance of developing new diagnostic approaches and useful tumor molecular markers for drug sensitivity to guide clinical chemotherapy regimens.
MicroRNAs (miRNAs) are a class of conserved endogenous small molecule non-protein-coding RNAs with sizes of 17–25 nucleotides. Which functional as post-transcription regulators and widely exists in the life progress of animals and plants (7). They can negatively regulate gene expression by perfect or nearly perfect complementarity to 3'untranslated region (3'-UTR) of protein-coding mRNA in a sequence specific manner, which induces mRNA degradation or protein translation repression (8). Bioinformatics database shows that one miRNA can target a variety of gene sequences, also one gene may also receive the control of multiple miRNAs. Those progress could be implicated in the regulation of almost every biological processes of organisms, especially in processes during tumorigenesis, such as proliferation, apoptosis, metastasis, and angiogenesis (9).
Mutations occur in pre-miRNA and mature miRNA, especially in the seed region, may result in deregulation of target gene expression by altering miRNAs expression or maturation (10-13), consequently contributing to cancer susceptibility, prognosis and chemotherapy sensitivity (14-17). Recently, an increasing number of researches have discussed the associations between miRNAs genetic variants and human cancers susceptibility, including lung cancer (18-21). However, the roles of genetic variants of miRNAs in platinum-based chemotherapy resistance in lung cancer are still unknown. Our study tried to verify the impact of pre-miRNA SNPs mutation on the lung cancer risk and explore the effect of miRNA mutation on platinum-based chemotherapy response.
Methods
Subjects
From November 2011 to August 2014, a total of 507 Chinese Han lung cancer patients were enrolled from the Affiliated Cancer Hospital and Xiangya Hospital of Central South University (Changsha, Hunan, China), and 215 healthy controls without any diseases physically examined in the Xiangya hospital were selected. Our study was approved by the Ethics Committee of Xiangya School of Medicine, Central South University and the registration numbers are CTXY-110008-2 and CTXY-110008-3. The clinical research admission was approved by Chinese Clinical Trial Registry with registration numbers of ChiCTR-RO-12002873 and ChiCTR-RCC-12002830, and written informed consent was obtained from all participants. The inclusion criteria were as follows: patients were histopathologically or cytologically diagnosed as lung cancer; patients were received at least two cycle of platinum-based chemotherapy. Exclusion criteria contain pregnancy or lactation, active infection, symptomatic brain or leptomeningeal metastases, and/or previous or concomitant malignancies.
A total of 386 patients treated with at least 2 cycle of platinum-based chemotherapy were selected in our Chemotherapy sensitivity analyses. According to the Response Evaluation Criteria (RECIST) guideline (version 1.1) for solid tumors (22), the selected objects were classified as complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). In our study, patients with CR or PR were designated platinum sensitivity, while PD or SD were regarded as platinum resistance.
DNA extraction, SNP selection and genotyping
Peripheral blood samples (5 mL) were obtained from each participating individuals and stored at −20 °C less than one week. The genomic DNA was isolated using a commercially available Genomic DNA Purification Kit (Wizard Genomic DNA Purification Kit, A1620; Promega, Madison, WI, USA) according to the standard protocols, the genomic DNA solution were stored at −20 °C orthonormal until use. For the target SNPs selection, two basic principles were considered. One was that, the minor allele frequency (MAF) larger than 5% in Chinese Han population. The other was that the SNPs might be associated with lung cancer susceptibility, prognosis or toxicity. To achieve these two criteria, we searched the published data concern about miRNA mutations and lung cancer. In addition, the databases of dbSNP (http://www.ncbi.nlm.nih.gov/snp), International HapMap Project (http://www.hapmap.org), and the online miRNA polymorphism databases (http://www.bioguo.org/miRNASNP/) were browsed. Eventually, 9 potentially functional SNPs were selected in this work.
All the polymorphisms were conducted by the Sequenom Mass Array Genotype Platform (Sequenom, San Diego, California, USA). Primers were designed in the soft of AssayDesigner (version 3.1).
Statistical analysis
The Hardy-Weinberg equilibrium analysis was carried out for the study participants using the χ2-test. The relationship of genotypes with lung cancer risk and chemotherapy response were examined by logistic regression analysis, the odds rations (OR) and their 95% confidence intervals (CI) were used to evaluate the results. And correction of potential confounder analysis was also made by adjusting for age, sex, smoking status, stage, histological type and chemotherapy regimens. Differences were considered statistically significant when two-tailed P value was less than 0.05. The statistical analyses above were performed by PLINK 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) and SPSS 18.0 for Windows (IBM, Inc., Chicago, IL, USA). The figures were performed using StataSE version 12 for Windows (StataCorp, CollegeStation, TX, USA).
Results
Demographic and clinical characteristics of the study population
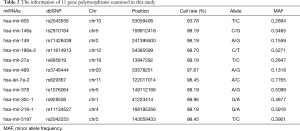
The genotyping was taken in a total of 507 lung cancer patients (398 males and 109 females) and 215 (84 males and 131 females) health controls. Among the lung cancer patients, only 386 patients, including 143 sensitive and 243 resistant individuals, received at least two cycles of platinum-based chemotherapy, and were enrolled in our study to investigate the role of miRNA polymorphisms in platinum-based chemotherapy response. The detailed information of clinical characteristics were summarized in Table 1. A significant difference was found in sex and histology between response and non-response (P=0.020 and 0.000, respectively). Other clinical characteristics, such as age, smoking status, stage, chemotherapy regimen were comparable in both groups. The information of 9 polymorphisms genotyped in this study were shown in Table 2, and the call rates of all the SNPs were above 95%, except rs2043556 with a call rate of 93.78%. The primer sequences for all the selected mutations were given in Table 3.

Full table
Full table

Full table
Association of functional miRNA polymorphisms with lung cancer susceptibility
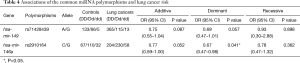
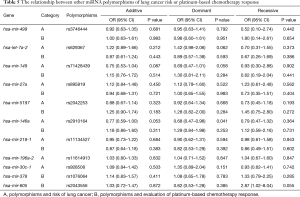
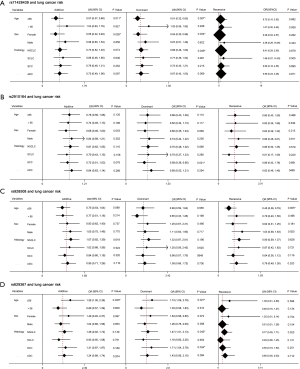
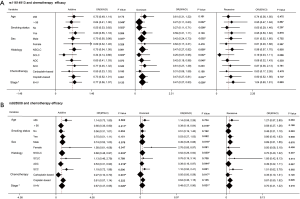
Preliminary analyses revealed that miR-149 rs71428439 and miR-146a rs2910164 were associate with the occurrence of lung cancer (P=0.042 and 0.022, respectively). As shown in Table 4, after adjusting age and sex, rs2910164 still has significantly association with lung cancer susceptibility in dominant model (OR=0.67, P=0.041) and has a marginal statistical effect in additive model (OR=0.77, P=0.052). While the rs71428439 have marginal statistical effect in dominate model with P value equal to 0.057. The other results were listed in Table 5 (mark A). In order to explore the deeper influence of miRNA mutation on the risk of lung cancer susceptibility, subgroup analyses were performed and the results (Figure 1) shown that rs71428439 was associated with NSCLC in dominant mode, the female patient or younger than 56 years subjects with this polymorphism may have lower risk of lung cancer in both additive and dominant modes. miR-146a rs2910164 polymorphism was associated with susceptibility of lung cancer of SCC patients in a dominant model. In subgroup under 57 years miR-30c-1 rs928508 polymorphism in recessive model and miR-let-7a-2 rs629367 polymorphism in dominant model were associated with lung cancer susceptibility (P=0.005 and 0.021, respectively). Let-7a-2 rs629367polymorphism was also associated with susceptibility of lung cancer of NSCLC and SCC individuals in a dominant model (P=0.043 and 0.034, respectively).
Full table
Full table
Effects of functional miRNA polymorphisms on the therapeutic efficacy of platinum-based chemotherapy in lung cancer patients
A total of 386 patients who received at least two cycles of platinum-based cure had been studied in the chemotherapy sensitivity analyses. After adjustment for age, sex, smoking status, stage, histology, and chemotherapeutic regimen, we found that carriers with T allele of miR-196a-2 rs11614913 were more resistance to platinum-based chemotherapy both in additive and dominant models (OR=0.67, 95% CI: 0.50–0.91, P=0.010 and OR=0.43, 95% CI: 0.26–0.72, P=0.001, respectively). And patients with A allele of miR-30c-1 rs928508 shown higher sensitivity to platinum-based chemotherapy in additive and dominant models (OR=0.69, 95% CI: 0.50–0.95, P=0.022 and OR=0.53, 95% CI: 0.31–0.90, P=0.018, respectively) (Table 6). The other results were listed in Table 5 (marked as B). In subgroup analyses, rs11614913 and rs928508 were affect chemotherapy response on certain level (summarized in Figure 2). For NSCLC, III and IV stage NSCLC, AC and limited-stage SCLC, the rs11614913 mutant individuals were more resistance to platinum-based chemotherapy in dominant model. We also found that in both additive and dominant models, rs11614913 polymorphism affected cisplatin-based chemotherapy response in male, SCLC, non-smoking, and over 56 years patients. MiR-30c-1 rs928508 was associated with chemotherapy response in additive model and dominant models for NSCLC, III IV stage of NSCLC, and AC subgroups, as well as in dominant model for male subgroup.

Full table
Discussion
In the current study, we investigated the association of 9 functional polymorphisms in pre-miRNAs on the risk of the lung cancer and the influence of platinum-based chemotherapy response in Chinese lung cancer patients. Our results showed that SNPs rs71428439 (miR-149), rs2910164 (miR-146a), rs629367 (let-7a-2) and rs928508 (miR-30c-1) might be related with the lung cancer prevalence, and polymorphisms of rs11614913 (miR-196a-2), rs9280508 (miR-30c-1) were significantly influence the response to platinum-based chemotherapy. To our knowledge, this is the first investigation about the association between functional polymorphisms of miRNAs and platinum-based chemotherapy response in Chinese lung cancer patients. These made it possible to do a rapid screening of lung cancer risk and drug resistance about platinum-based chemotherapy in lung cancer parents without any invasive examination, only blood was needed.
MiRNAs are a class of small non-coding RNA molecules, play important roles in many biological and physiological processes, aberrant microRNA expression has been found in several diseases including lung cancer (23-28). MiR-149 is down regulated in several cancer types (29,30) and is believed to act as a versatile tumor-suppression miRNA by regulating the expression of many genes such as AKT1 (31), GIT1 (32), ZBTB2 (33), FOXM1 (34). Recently studies showed that miR-149 rs71428439 polymorphism was significantly associated with increased clear cell renal cell carcinoma (CCRCC) and hepatocellular carcinoma (HCC) risk (28,35). We didn’t find the association of this polymorphism with lung cancer risk in this study, but in NSCLC, female, age under 57 years subgroups, carriers with A allele of this mutation has a higher risk of lung cancer. Ding et al. observed that miR-149 can conquer mitochondrial related apoptosis through targeting the pro-apoptotic protein Puma, meanwhile the A allelic miR-149 precursor made a higher production of mature miR-149 (36), another study found that GG genotype expressed lower level of miRNA-149 compared with AA and AG genotypes in HCC tissues (35). All these data suggested that miRNA-149 A allele carriers may be more vulnerable carcinogenesis. Shi et al. (37) report that decreasing miR-146a is associated with the aggressiveness of human oral squamous cell carcinoma. Another research indicate that miR-146a expression levels were lower in lung cancer cells and could regulate COX-2 expression involved in the development of a metastatic condition (38). A functional mutation in miR-146a rs2910164 may act as a useful molecular marker for cardiovascular disease (39,40), and also for various cancer types (19,41,42). For Korean population rs2910164 may contribute to genetic susceptibility to lung cancer (43), it also can be used as a prognostic marker for NSCLC patients with surgically resected at early-stage in Chinese people (44) and another study revealed that rs2910164 CG genotype group had a higher risk of NSCLC than GG genotype group (45). Our results confirmed that polymorphism rs2910164 associated with the risk of lung cancer, the CC genotype and C allele distribution in the lung patient were significantly higher than that of the controls. As previous studies have found that rs2910164 G/C polymorphism affects both the efficiency of pri-miRNA processing and protein binding to the pre-miRNA product of this reaction (46), and miR-146a CC genotypes have a lower expression level compare to GG/GC genotypes subgroup in tumor tissues (47), which may partly explain the tendency that rs2910164 C allele mutation subjects had a higher risk of lung cancer in our study.
Let-7a is a member of the family of let-7 contains of let-7a-1/2/3, supposed to have antitumor effect in lung cancer (48). Guan et al. reported that 1,25-(OH)2VD3 could up-regulate the transcription of let-7a-2 in lung cancer cells, which might mediate the anti-proliferation effects in lung cancer cells (49). Recently Xu et al. reported that a polymorphism rs629367 could affect the mature of let-7a, further associated with gastric cancer risk and survival (50). In this study, we founded that rs629367 C allele could increase the risk of lung cancer in age no more than 56 years subgroup.
Researches shown that the expression level of miR-30c was lower in many cancers, including lung cancer, which affect the process of epithelial mesenchymal transition (51,52). Fang et al. made an evidence that miR-30c could promote the sensibility to doxorubicin in breast cancer cell by regulated p38 mitogen-activated protein kinase (p38MAPK) pathway (53). The genomics research revealed that genetic polymorphism rs928508 could influence the gastric cancer risk and the prognostic of NSCLC (54,55). In the present study we found that this polymorphism may associate with lung cancer risk in age under 57 years old person. Here, for the first time we found that the mutation of this SNP would increase platinum-based chemotherapy sensitivity in lung cancer. The subgroup analyses also made a same impact in NSCLC, III and IV stage NSCLC, AC, male subgroups. Previous study had testified that themiR-30c AA genotype had a higher expression than AG/GG genotypes (56), which given evidence to the chemotherapy sensitive analyses. As miRNAs could bind to various genes, including oncogenes and tumor suppressor genes, other targets of miR-30c needed to be figure out, which would give us a better understanding of the influence of rs928508 on chemotherapy response.
MiR-196a encoded at two paralogous locations in the B and C mammalian HOX clusters, contains miR-196a-1 located on chromosome 17 (17q21.32) and miR-196a-2 located on chromosome 12 (12q13.13), taken part in regulating cell differentiation (57,58). Accumulating number of studies showed miR-196a is highly expressed in various tumor tissues, including esophageal carcinoma, gastric carcinoma, pancreatic cancer (59) and lung cancer (45). Study shows that inhibition of miR-196a could reverse cisplatin resistance of A549/DDP cell lines (60). Recently a functional SNP in miR-196a-2 rs11614913 was researched. Shen et al. found that miR-196a2 rs11614913was associated with an increased esophageal squamous cell carcinoma (ESCC) risk in a Chinese population (61). Deng et al. revealed this polymorphism was associated with a decreased risk of bladder cancer (42). Unfortunately we didn’t reveal the association between rs11614913 and lung cancer risk, however our study was the first to reveal that polymorphism rs11614913 was significantly influence platinum-based chemotherapy response in lung cancer patients. Another study confirmed that rs11614913 was associated with severe toxicity after platinum-based regimen in advanced NSCLC (62). Analysis of mature miRNA expression confirmed that carriers with T allele in miR-196a-2 rs11614913 dramatically inhibited production of their mature products (63), while Vinci and colleagues found that rs11614913 CC genotype significant associated with high expression of miR-196a-2 (45). Thereby the alteration of miR-196a-2 expression may involve in cisplatin efficacy. On the other hand, the mutation may also influence genes binding effects (64), which confounding the drug resistance.
The data presented here suggested that miR-149 rs71428439, miR-146a rs2910164, let-7a-2 rs629367 and miR-30c-1 rs928508 were significantly associated with lung cancer susceptibility, what’s more miR-30c-1 rs928508 and miR-96a-2 rs11614913 were significantly related to platinum-based chemotherapy response. It is conceivable that those genetic polymorphisms may be useful in predicting the occurrence risk of lung cancer and platinum-based chemotherapy response in lung cancer patients.
It should not be ignored that the current study had some limitations. On the one hand, after the multiple testing of False Discovery Rate (FDR) our results didn’t make a statistical difference. On the other hand, an independent validation for these SNPs needs to be arranged to get more credible conclusion. After that, other in depth mechanism research needs to be carried out to figure out how these SNPs influence lung cancer risk and chemotherapy response.
Acknowledgements
We thank all patients who participated in the study.
Funding: This work was supported by the National High-tech R&D Program of China (863 Program) (2012AA02A517), National Natural Science Foundation of China (81373490, 81573508, 81573463), and Hunan Provincial Science and Technology Plan of China (2015TP1043).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics Committee of Xiangya School of Medicine, Central South University and the registration numbers are CTXY-110008-2 and CTXY-110008-3. The clinical research admission was approved by Chinese Clinical Trial Registry with registration numbers of ChiCTR-RO-12002873 and ChiCTR-RCC-12002830. Written informed consent was obtained from all participants.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Rothschild SI. Epigenetic Therapy in Lung Cancer - Role of microRNAs. Front Oncol 2013;3:158. [Crossref] [PubMed]
- Nilssen Y, Strand TE, Fjellbirkeland L, et al. Lung cancer survival in Norway, 1997-2011: from nihilism to optimism. Eur Respir J 2016;47:275-87. [Crossref] [PubMed]
- Li XP, Yin JY, Wang Y, et al. The ATP7B genetic polymorphisms predict clinical outcome to platinum-based chemotherapy in lung cancer patients. Tumour Biol 2014;35:8259-65. [Crossref] [PubMed]
- Xu X, Han L, Duan L, et al. Association between eIF3alpha polymorphism and severe toxicity caused by platinum-based chemotherapy in non-small cell lung cancer patients. Br J Clin Pharmacol 2013;75:516-23. [Crossref] [PubMed]
- Chen J, Yin JY, Li XP, et al. Association of Wnt-Inducible Signaling Pathway Protein 1 Genetic Polymorphisms With Lung Cancer Susceptibility and Platinum-Based Chemotherapy Response. Clin Lung Cancer 2015;16:298-304.e1-2.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537-61. [Crossref] [PubMed]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389-402. [Crossref] [PubMed]
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 2007;16:1124-31. [Crossref] [PubMed]
- Zorc M, Skok DJ, Godnic I, et al. Catalog of microRNA seed polymorphisms in vertebrates. PLoS One 2012;7. [Crossref] [PubMed]
- Rotunno M, Zhao Y, Bergen AW, et al. Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br J Cancer 2010;103:1870-4. [Crossref] [PubMed]
- Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 2008;118:2600-8. [PubMed]
- Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev 2009;18:1183-7. [Crossref] [PubMed]
- Pardini B, Rosa F, Naccarati A, et al. Polymorphisms in microRNA genes as predictors of clinical outcomes in colorectal cancer patients. Carcinogenesis 2015;36:82-6. [Crossref] [PubMed]
- Geng JQ, Wang XC, Li LF, et al. MicroRNA-related single-nucleotide polymorphism of XPO5 is strongly correlated with the prognosis and chemotherapy response in advanced non-small-cell lung cancer patients. Tumour Biol 2016;37:2257-65. [Crossref] [PubMed]
- Wei Y, Li L, Gao J. The association between two common polymorphisms (miR-146a rs2910164 and miR-196a2 rs11614913) and susceptibility to gastric cancer: A meta-analysis. Cancer Biomark 2015;15:235-48. [Crossref] [PubMed]
- Upadhyaya A, Smith RA, Chacon-Cortes D, et al. Association of the microRNA-Single Nucleotide Polymorphism rs2910164 in miR146a with sporadic breast cancer susceptibility: A case control study. Gene 2016;576:256-60. [Crossref] [PubMed]
- Zhang MW, Jin MJ, Yu YX, et al. Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol Carcinog 2012;51 Suppl 1:E21-31. [Crossref] [PubMed]
- Nikolic Z, Savic Pavicevic D, Vucic N, et al. Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp Mol Pathol 2015;99:145-50. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Jin Y, Liu Y, Zhang J, et al. The Expression of miR-375 Is Associated with Carcinogenesis in Three Subtypes of Lung Cancer. PLoS One 2015;10. [Crossref] [PubMed]
- Zhao W, Zhao JJ, Zhang L, et al. Serum miR-21 level: a potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med 2015;8:14759-63. [PubMed]
- Zhang F, Yang R, Zhang G, et al. Anticancer function of alpha-solanine in lung adenocarcinoma cells by inducing microRNA-138 expression. Tumour Biol 2016;37:6437-46. [Crossref] [PubMed]
- Gu R, Huang S, Huang W, et al. MicroRNA-17 family as novel biomarkers for cancer diagnosis: a meta-analysis based on 19 articles. Tumour Biol 2016;37:6403-11. [Crossref] [PubMed]
- Tian Y, Wei W, Li L, et al. Down-Regulation of miR-148a Promotes Metastasis by DNA Methylation and is Associated with Prognosis of Skin Cancer by Targeting TGIF2. Med Sci Monit 2015;21:3798-805. [Crossref] [PubMed]
- Wang Z, Wei M, Ren Y, et al. miR149 rs71428439 polymorphism and risk of clear cell renal cell carcinoma: a case-control study. Tumour Biol 2014;35:12127-30. [Crossref] [PubMed]
- Xue L, Wang Y, Yue S, et al. Low MiR-149 expression is associated with unfavorable prognosis and enhanced Akt/mTOR signaling in glioma. Int J Clin Exp Pathol 2015;8:11178-84. [PubMed]
- Molina-Pinelo S, Gutierrez G, Pastor MD, et al. MicroRNA-dependent regulation of transcription in non-small cell lung cancer. PLoS One 2014;9. [Crossref] [PubMed]
- Zhang Y, Guo X, Xiong L, et al. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol Cancer 2014;13:253. [Crossref] [PubMed]
- Chan SH, Huang WC, Chang JW, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene 2014;33:4496-507. [Crossref] [PubMed]
- Wang Y, Zheng X, Zhang Z, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One 2012;7. [Crossref] [PubMed]
- Ke Y, Zhao W, Xiong J, et al. miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochem Res Int 2013;2013. [Crossref] [PubMed]
- Wu J, Lv S, An J, et al. Pre-miR-149 rs71428439 polymorphism is associated with increased cancer risk and AKT1/cyclinD1 signaling in hepatocellular carcinoma. Int J Clin Exp Med 2015;8:13628-33. [PubMed]
- Ding SL, Wang JX, Jiao JQ, et al. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. J Biol Chem 2013;288:26865-77. [Crossref] [PubMed]
- Shi Z, Johnson JJ, Jiang R, et al. Decrease of miR-146a is associated with the aggressiveness of human oral squamous cell carcinoma. Arch Oral Biol 2015;60:1416-27. [Crossref] [PubMed]
- Cornett AL, Lutz CS. Regulation of COX-2 expression by miR-146a in lung cancer cells. Rna 2014;20:1419-30. [Crossref] [PubMed]
- Huang S, Lv Z, Deng Q, et al. A Genetic Variant in Pre-miR-146a (rs2910164 C>G) Is Associated with the Decreased Risk of Acute Coronary Syndrome in a Chinese Population. Tohoku J Exp Med 2015;237:227-33. [Crossref] [PubMed]
- Zhu J, Yue H, Qiao C, et al. Association Between Single-Nucleotide Polymorphism (SNP) in miR-146a, miR-196a2, and miR-499 and Risk of Ischemic Stroke: A Meta-Analysis. Med Sci Monit 2015;21:3658-63. [Crossref] [PubMed]
- Chen J, Cao X, Zhang H. MiR-146a rs2910164 polymorphism is associated with hepatocellular carcinoma: a meta-analysis. Int J Clin Exp Med 2015;8:15852-6. [PubMed]
- Deng S, Wang W, Li X, et al. Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J Surg Oncol 2015;13:297. [Crossref] [PubMed]
- Jeon HS, Lee YH, Lee SY, et al. A common polymorphism in pre-microRNA-146a is associated with lung cancer risk in a Korean population. Gene 2014;534:66-71. [Crossref] [PubMed]
- Hong MJ, Choi YY, Jang JA, et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. J Thorac Oncol 2013;8:703-10. [Crossref] [PubMed]
- Vinci S, Gelmini S, Pratesi N, et al. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med 2011;49:2073-80. [Crossref] [PubMed]
- Jazdzewski K, Murray EL, Franssila K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A 2008;105:7269-74. [Crossref] [PubMed]
- Jia Y, Zang A, Shang Y, et al. MicroRNA-146a rs2910164 polymorphism is associated with susceptibility to non-small cell lung cancer in the Chinese population. Med Oncol 2014;31:194. [Crossref] [PubMed]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007;67:7713-22. [Crossref] [PubMed]
- Guan H, Liu C, Chen Z, et al. 1,25-Dihydroxyvitamin D3 up-regulates expression of hsa-let-7a-2 through the interaction of VDR/VDRE in human lung cancer A549 cells. Gene 2013;522:142-6. [Crossref] [PubMed]
- Xu Q, Dong Q, He C, et al. A new polymorphism biomarker rs629367 associated with increased risk and poor survival of gastric cancer in chinese by up-regulated miRNA-let-7a expression. PLoS One 2014;9. [Crossref] [PubMed]
- Irani S, Hussain MM. Role of microRNA-30c in lipid metabolism, adipogenesis, cardiac remodeling and cancer. Curr Opin Lipidol 2015;26:139-46. [Crossref] [PubMed]
- Zhong Z, Xia Y, Wang P, et al. Low expression of microRNA-30c promotes invasion by inducing epithelial mesenchymal transition in non-small cell lung cancer. Mol Med Rep 2014;10:2575-9. [Crossref] [PubMed]
- Fang Y, Shen H, Cao Y, et al. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res 2014;47:60-9. [Crossref] [PubMed]
- Mu YP, Su XL. Polymorphism in pre-miR-30c contributes to gastric cancer risk in a Chinese population. Med Oncol 2012;29:1723-32. [Crossref] [PubMed]
- Hu Z, Shu Y, Chen Y, et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am J Respir Crit Care Med 2011;183:641-8. [Crossref] [PubMed]
- Chen JP, Liu Y, Hu ZB, et al. Zhonghua Zhong Liu Za Zhi 2012;34:664-8. [Single nucleotide polymorphism in flanking region of miR-30c influences the maturing process of miR-30c in lung carcinoma]. [PubMed]
- Qiu R, Liu Y, Wu JY, et al. Misexpression of miR-196a induces eye anomaly in Xenopus laevis. Brain Res Bull 2009;79:26-31. [Crossref] [PubMed]
- Kim YJ, Bae SW, Yu SS, et al. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res 2009;24:816-25. [Crossref] [PubMed]
- Chen C, Zhang Y, Zhang L, et al. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med 2011;15:14-23. [Crossref] [PubMed]
- Li JH, Luo N, Zhong MZ, et al. Inhibition of microRNA-196a might reverse cisplatin resistance of A549/DDP non-small-cell lung cancer cell line. Tumour Biol 2016;37:2387-94. [Crossref] [PubMed]
- Shen F, Chen J, Guo S, et al. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol 2016;37:4777-84. [Crossref] [PubMed]
- Zhan X, Wu W, Han B, et al. Hsa-miR-196a2 functional SNP is associated with severe toxicity after platinum-based chemotherapy of advanced nonsmall cell lung cancer patients in a Chinese population. J Clin Lab Anal 2012;26:441-6. [Crossref] [PubMed]
- Qi P, Wang L, Zhou B, et al. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res 2015;14:6289-96. [Crossref] [PubMed]
- Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res 2009;69:5970-7. [Crossref] [PubMed]


