What information could the main actors of liquid biopsy provide? —a representative case of non-small cell lung cancer (NSCLC)
Introduction
Recent randomized studies showed that epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) significantly improved the progression-free survival (PFS) of advanced non-small cell lung cancer (NSCLC) harboring activating EGFR mutations, compared to chemotherapy (1-3). Conversely, TKIs do not offer any benefit to EGFR wild-type patients (1).
Collecting proper tissue samples is the key issue for molecular analyses, although it is not always an easy task, particularly in advanced NSCLC (4). Therefore, a blood draw could provide an alternative source of tumor samples.
In NSCLC, circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), simultaneously present in patients’ blood, could broaden, in concert, our knowledge on tumor characteristics and behavior. In particular, these biomarkers can be useful to detect “druggable” mutations and monitor TKIs, thus improving the prognosis (5).
CTCs can form metastases and, in NSCLC, they express both epithelial and mesenchymal markers (6). Regarding cell-free DNA (cfDNA), its concentration and/or tracking specific tumor mutations (ctDNA) could be used to monitor treatment effectiveness (7).
To obtain a more comprehensive view of the circulating cells, we investigated both epithelial cell adhesion molecule (EpCAM)-positive and EpCAM-low/negative cells, and evaluated in both subsets 2 cytokeratin-profiles. By monitoring CTCs and ctDNA at multiple time points throughout the care, we could reveal important information on disease evolution.
Through a single-tube procedure, this work demonstrated how and when these actors of liquid biopsy could be useful to improve the clinical management of the patient.
Materials and Methods
Blood samples
Blood samples were collected upon informed consent (IOV-IRCCS study “Individualized treatment of patients with advanced NSCLC: potential application for CTCs molecular and phenotypical profiling”, NCT02407327).
We collected serial blood samples at baseline, at the end of the first and second cycle of therapy, after radiological assessment, after surgery before starting the new treatment and after 6 months of post-surgery adjuvant treatment.
At each time point, we performed the quantification of EpCAM-positive and EpCAM-low/negative cells. Furthermore, we analyzed ctcDNA and cfDNA by droplet digital PCR (ddPCR) to disclose the allele frequency of EGFR L858R and T790M.
Detection of EpCAM-positive and EpCAM-low/negative CTCs
We enumerated EpCAM-positive CTCs in whole blood with the CellSearch System (CS) (Menarini, Italy) according to the manufacturer’s instructions and user’s guidelines [standard assay (SA)] (8).
Furthermore, to investigate the full cytokeratin (CK) profile of NSCLC CTCs, we integrated the CTC assay, as previously described, using the CXC Kit (Menarini, Italy) with FITC-conjugated anti-CK7, -CK14 (clone LP5K and LL002, respectively, Millipore, Billerica MA, USA), and anti-C11 (clone C11, ACZON, Bologna, Italy) [expanded assay (EA)] (9).
EpCAM-low/negative cells were collected downstream of the CellSearch processing, using a 5 µm-pore microsieve membrane (VyCAP, Deventer, The Netherlands), as previously described (10).
The cells on microsieve were stained using a cocktail of fluorescently labelled antibodies, namely: FITC-conjugated detecting CK1-10, 13–16, 18, and 19 (AE1/AE3, eBioscience; C11, ACZON), APC-conjugated anti-CD16 and -CD45 (Invitrogen), and DAPI for nuclear staining (SA). The EA also included CK7 and 14 (Millipore) antibodies. Cells were scored as DAPI+CK+CD45−CD16−, with a minimum cell size of 4 µm × 4 µm, and a nucleus occupying at least 50% of the CK+ cytoplasm.
DNA whole genome amplification (WGA) from CTCs
The content of the CS cartridge was subjected to WGA with Ampli1-WGA Kit (Menarini Silicon Biosytems, BO, Italy) according to the manufacturer’s instructions. Similarly, EpCAM-low/negative cell DNA was extracted using the QIAmp DNA Micro Kit (QIAGEN, Hilden, Germany), following the manufacturer’s protocol, and subjected to WGA.
Circulating cfDNA
The EDTA blood samples were centrifuged at 1,600 g for 10 minutes, followed by 16,000 g for another 10 minutes, and the plasma supernatant was stored at −80 °C until analysed. Plasma DNA was purified with QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA, USA), as recommended by the manufacturer. The extracted DNA was stored at −80 °C.
ddPCR assays
Allele frequency of EGFR L858R and T790M was measured with the QX200 droplet digital PCR system by the commercial PrimePCR ddPCR mutation assays. The data was acquired and analysed by QuantaSoft analysis software version 1.7.4 (BioRad, Hercules, CA, USA).
Results
Clinical history
In November 2015, a 65-year-old male, former smoker came to our unit reporting chest pain and exertional dyspnea.
The patient underwent a CT scan showing a lung mass of 5.5 cm located in the upper left lung (Figure 1A); the neighboring mediastinal lymph nodes were involved. We performed a bronchoscopy, with bronchial biopsies. The immunohistochemistry revealed a P63-negative, TTF-1-positive adenocarcinoma. The mutation status in bronchial specimen was EGFR and ALK negative.
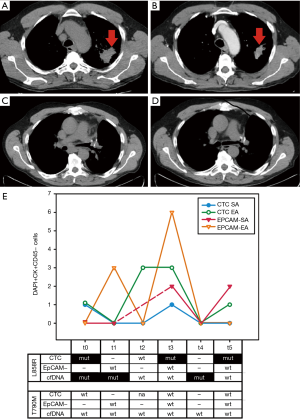
Based on this, we planned neo-adjuvant treatment (cis-platinum + vinorelbine). The patient showed a partial response (PR) according to RECIST 1.1 criteria (11) (Figure 1B), and then underwent surgical lobectomy. The surgical sample showed EGFR exon 21 mutation (L858R).
According to NCCN guidelines, the patient received three cycles of cisplatin and vinorelbine, as adjuvant treatment, until May 2016 (12).
In August 2016 however, an FDG-PET-CT documented disease progression in perihilar region of left lung (Figure 1C).
The patient began a second-generation TKI treatment (afatinib 40 mg/day). Two months later, the magnetic resonance imaging (MRI) documented brain metastasis that we treated with stereotactic radiotherapy, along with mediastinum radiotherapy. According to the last FDG-PET-CT, in February 2017, the patient showed stable disease (SD) (Figure 1D), only paratracheal lymph nodes being metabolically active.
Table 1 compares clinical-pathological characteristics, treatments and imaging reports throughout the continuum of the care, with findings obtained through liquid biopsy.
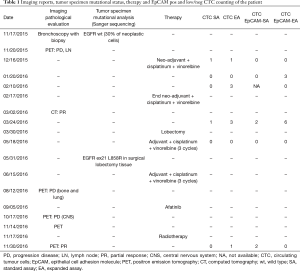
Full table
EpCAM-positive CTCs, EpCAM-low/negative CTCs and ctDNA
Concerning CTCs, we noticed a saw-toothed count pattern in longitudinal graphs for all four subpopulations, EpCAM-positive SA and EA and EpCAM-low/negative EA and SA (Figure 1E). At baseline (t0), we only found EpCAM-positive CTCs (both EA and SA), while the EpCAM-low/negative subpopulation emerged after the first cycle of chemotherapy, when the EpCAM-positive subpopulation disappeared (t1); interestingly, after the next cycle of therapy, the EpCAM-positive EA subpopulation reappeared (t2).
Notably, we highlighted the highest level in all CTC subgroups immediately before lobectomy (t3), while they were undetectable after lobectomy and 3 cycles of adjuvant + cis-platinum + vinorelbine (t4). Simultaneously, we analyzed by ddPCR the presence of EGFR mutated ctDNA at every time point, and DNA of CTC-positive samples, both EpCAM-positive and EpCAM-low/negative.
We found L858R positive ctDNA at baseline (t0), after two cycles of therapy (t1) and at lobectomy (t4).
Interestingly, the mutational profile of CTCs also changed during treatment: at baseline (t0), in the EpCAM-positive fraction, we found the EGFR L858R mutation that was undetectable in the biopsy specimen using Sanger Sequencing. We documented the same mutation in EpCAM-positive EA CTCs after two cycles of therapy (t3) and at the last point of observation (t5). Conversely, EpCAM-low/negative CTCs were always negative for L858R. The CTC and ctDNA samples analyzed were all negative for the T790M mutation.
Curiously, when CT showed a PR (Table 1 and Figure 1B,D), we noticed the presence of at least one CTC subpopulation; moreover, EpCAM-positive and EpCAM-low/negative CTCs persisted at the last time point (t5).
Discussion
The main challenge of targeted therapies in NSCLC, as well as in other malignancies, is tracking the disease dynamics to measure the effectiveness of treatment and to anticipate a change of therapy, when appropriate. Here, we demonstrated, through serial sampling of an individual patient, how the analysis of the different components of liquid biopsy could shed light on the tumor strategy to escape treatment.
Indeed, during platinum-based chemotherapy, the tumor releases EpCAM-low/negative CTCs; consistently with previous reports in others malignancies, where this treatment seems to recruit EMT-like CTCs (13,14).
However, the EpCAM-positive subpopulation reappeared at the end of the second cycle of therapy; this could imply an escape strategy of the disease and the coexistence of different CTC subpopulations.
Interestingly, at the last time point, when radiological control referred SD, we observed EpCAM-positive and EpCAM-low/negative cells, and found the L858R mutation in EpCAM-positive CTCs. On the one hand, we should not underestimate the presence of different subpopulations of CTCs in conjunction with a PR, especially if we consider that these cells represent an intermediate step toward metastasis. On the other hand, the fact that at least one of them is carrying an activating EGFR mutation demonstrates that TKIs are of potential benefit in this patient, as documented by the long-term control of his disease that we obtained with afatinib, from February 2017 until now. A consensus exists that CTCs are heterogeneous and could allow real-time tracking of the tumor transient states. Moreover, CTCs could show the spectrum of mutations and/or protein expression alterations present in cancer. Indeed, we noted here that the four CTC subpopulations allow the monitoring of the different steps of cancer evolution in response to treatment.
Furthermore, we observed a slight discordance about the L858R detection timing between the golden standard analysis of tumor specimens, CTCs and ctDNA.
Discrepancy between mutational status as documented in bronchial biopsies and surgical sample after lobectomy, is not surprising and we can easily explain it, because of a limited sampling offered by bronchial biopsies. Indeed, at this regard, cfDNA, intended as the sum of different neoplastic lesions present at any time in individual patient, seems to be more advantageous, when highly sensitive technique has been using to detect L858R, as ddPCR in our case report. At this regard, we should reflect about the superiority of ctDNA in choosing target treatment, also when bronchial specimens give negative results.
However, our findings do not support the superiority of ctDNA at any time of patient’ monitoring. For example, before lobectomy (t3), when likely chemotherapy affected the release of CTCs in the blood stream, we could detect EGFR mutation in EpCAM-positive tumor cells, but not in ctDNA. Indeed, it is conceivable that changing level of few cells, did not affect the total amount of ctDNA to result in EGFR detection by using this technique, whereas ctcDNA investigation was more useful at that time.
This finding suggests the complementary use of these tests (15): taking them together, in this patient they better reflected spatial and temporal heterogeneity of the tumor under the selective pressure of treatment.
To date, the development of resistance in cancer cells, acquired via clonal evolution during treatment, represents the main challenge of a successful targeted therapy. To bypass this bottleneck, an element of the utmost importance is an accurate design of the therapeutic strategy. However, to achieve this goal, we need robust biomarkers for serial tumor profiling (16). Indeed, high effort has been made, so far, to standardize and validate, in clinic, consensus analyses of liquid biopsy, as CTCs and ctDNA/miRNA, by several scientific consortia [e.g., STALKLUNG01, AIR Study Consortium (17), or CANCER_ID, www/cancer-id/eu], so that we are confident to receive soon advanced remarks at this regard.
In our study, the actors of liquid biopsy and their specific characteristics disclosed a polyclonal resistance that could allow up-front, combination therapy to prolong treatment response.
It still has to be clarified, in further larger studies, how the CTC subgroups join forces in high-risk patients. In the future, we might use this information to design a tailored therapeutic strategy to hinder the continuous tumor evolution.
Acknowledgements
Funding: This work was supported in part by grants from: (I) the Italian Ministry of Health, Proposal No: # GR-2010-2303193A, “Individualized treatments of patients with advanced NSCLC: potential application for CTCs molecular and phenotypical profiling” (PI: E Rossi); (II) M Manicone and MC Scaini were supported by post-doc fellowships funded by Innovative Medicine Initiative Joint Undertaking [115749] CANCER-ID (PI: R Zamarchi).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Informed Consent: Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
References
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Coghlin CL, Smith LJ, Bakar S, et al. Quantitative analysis of tumor in bronchial biopsy specimens. J Thorac Oncol 2010;5:448-52. [Crossref] [PubMed]
- Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-27. [Crossref] [PubMed]
- Soltermann A, Tischler V, Arbogast S, et al. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res 2008;14:7430-7. [Crossref] [PubMed]
- Tie J, Gibbs P. Sequencing Circulating Cell-Free DNA: The Potential to Refine Precision Cancer Medicine. Clin Chem 2016;62:796-8. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Rossi E, Basso U, Celadin R, et al. M30 neoepitope expression in epithelial cancer: quantification of apoptosis in circulating tumor cells by CellSearch analysis. Clin Cancer Res 2010;16:5233-43. [Crossref] [PubMed]
- de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015;5:12270. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol 2006;7:719-27. [Crossref] [PubMed]
- Chebouti I, Kasimir-Bauer S, Buderath P, et al. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget 2017;8:48820-31. [Crossref] [PubMed]
- Pavese JM, Bergan RC. Circulating tumor cells exhibit a biologically aggressive cancer phenotype accompanied by selective resistance to chemotherapy. Cancer Lett 2014;352:179-86. [Crossref] [PubMed]
- Madic J, Kiialainen A, Bidard FC, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer 2015;136:2158-65. [Crossref] [PubMed]
- Lopez JS, Banerji U. Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat Rev Clin Oncol 2017;14:57-66. [Crossref] [PubMed]
- Ilie M, Hofman V, Leroy S, et al. Use of circulating tumor cells in prospective clinical trials for NSCLC patients - standardization of the pre-analytical conditions. Clin Chem Lab Med 2018;56:980-9. [Crossref] [PubMed]


