ctDNA assessment of EGFR mutation status in Chinese patients with advanced non-small cell lung cancer in real-world setting
Introduction
For many decades, lung cancer has been the most common cancer and the leading cause of cancer death worldwide (1). Approximately 733 million new lung cancer cases have been estimated to occur in China each year (2). About 85% of lung cancers are non-small cell lung cancer (NSCLC) and approximately 10% of US, 13% of European, and 35% of East Asia patients harbor epidermal growth factor receptor (EGFR) mutations. EGFR-tyrosine kinase inhibitors (EGFR-TKIs) are highly effective clinical therapies for NSCLC patients with EGFR mutations which have showed an objective response rate (ORR) of almost 70% and prolonged progression-free survival (PFS) to 8–13 months (3-8). Previously both clinical trials and real-world settings have demonstrated the predictive and prognostic value of tissue-based EGFR mutation on patients receiving TKIs. Tissue-based genotyping has always been the gold standard test for detection of EGFR mutation (9). Yet, in at least 20% of patients especially after chemotherapy, tissue-based detection is not available for various reasons including insufficient availability of neoplastic tissue, lack of fitness of the available tissue for a biopsy or that a biopsy is not technically feasible (10). The circulating free tumor-derived DNA (ctDNA)-based mutation detection has shown to be very promising due to several significant advantages, including non-invasiveness, accessibility and the potential for repeated sampling. Several studies have demonstrated that it is feasible to assess EGFR mutation status by using ctDNA, which can be isolated from the plasma or serum of patients with NSCLC (11,12). As ctDNA analysis is technically challenging, it is important that the accuracy, suitability, and feasibility of use of ctDNA for mutation analysis in clinical practice are established. Many clinical centers have investigated the diagnostic accuracy of ctDNA for detection of EGFR mutation (13-15). The concordance rate of EGFR mutation between ctDNA and tumor tissues is largely dependent on detection techniques, and varies from 66% to 100% (16). In Caucasian population, the open-label IFUM study found EGFR mutation status concordance was 94% in patients with EGFR mutation-positive NSCLC between 652 evaluable matched tissue/cytologic and plasma (ctDNA) samples; in European and Japanese population, the ASSESS study evaluated the prevalence and accuracy of ctDNA EGFR mutation (12). The overall EGFR mutation frequency was 9% for evaluable plasma samples (8% for European and 13% for Japanese). Overall concordance of mutation status was 89% (sensitivity 46%, specificity 97%, PPV 78%, and NPV 90%). Although several studies have evaluated the prevalence of ctDNA EGFR mutation, while that in Chinese population is still unknown. EGFR mutation in plasma ctDNA by amplification refractory mutation system (ARMS) has been widely used in clinical settings in China. However, the prevalence of EGFR mutations in ctDNA was still unknown in the real world. This large-scale study (NCT02623257) aimed to explore the prevalence of EGFR mutations and determine the correlation of EGFR mutation status with clinical characteristics.
Methods
Study population
This observational, multi-institution, large-scale diagnostic study was performed between January 2016 and July 2017. The protocol was approved by the Institutional Review Board of Hangzhou First People’s Hospital (No. HZFH 2015-47-01). All patients signed the informed consent. This study had been registered on clinicaltrials.gov (NCT02623257). All methods were performed in accordance with the relevant guidelines and regulations. Patients were considered eligible and enrolled in this study if they met the following criteria: (I) histologically confirmed as stage III/IV NSCLC; (II) received ≤1st line chemotherapy; (III) with samples collected more than 2 weeks after 1st line chemotherapy; (IV) with samples collected before any following treatment; (V) with collected data for clinical features and follow-up treatment.
Sample collection and DNA extraction
The 10–15 mL of peripheral blood was collected in a cell-free DNA protection vacuum tube (AmoyDx, Xiamen, China), which contains cell-free DNA protection reagent to keep DNA stable within 7 days at 4–25 °C. The blood samples were transported to Center for Translational Medicine of Hangzhou First People’s Hospital within 36 hours for further processing. For DNA extraction, the blood samples were centrifuged at 2,500 g for 10 minutes at 4 °C. The supernatant was transferred to a new tube and centrifuged at 15,800 ×g for 15 minutes at 4 °C. The plasma supernatant was stored at −80 °C. The cell free DNA from 1.5 mL plasma was extracted with QIAamp Circulating Nucleic Acid kit according to the manufacturer’s instructions (CAS# 55114, Qiagen, Hilden, Germany).
Detection of EGFR mutations in plasma ctDNA by ARMS
EGFR mutations in plasma ctDNA were determined by using ADx-ARMS (amplification refractory mutation system) kit (Amoy Diagnostics, Xiamen, China), and all experiments and genotyping calling were performed according to the manufacturer’s instructions (17).
Statistical analysis
Continuous data of normal distribution between two groups were compared using Student’s t test, Wilcoxon two sample tests were performed when continuous data did not follow the normal distribution. Categorical data between the two groups were compared using the Chi-square or Fisher’s exact test. Unconditional multiple logistic regression were performed to estimate the risk factors of EGFR mutation, independent variables included age, gender, smoking, chemotherapy, and pathology of tumor. Significance in all the analyses was assessed at 0.05 levels. All tests were performed with two-tailed. The analyses were performed using SAS software (version 9.3, SAS Institute, Cary, NC, USA).
Results
Patient characteristics
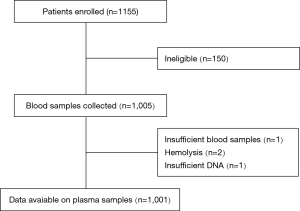
A thousand one hundred and fifty-five patients with advanced or recurrent NSCLC between January 2016 and July 2017 were enrolled consecutively. Excluding 154 ineligible patients, 1,001 were included in analysis. The median age of the patients was 67 years (19–94 years). 60.5% of patients were male. 55.6% had no smoking history. The type of NSCLC was predominantly adenocarcinoma (86.7%) and 65.9% of patients were diagnosed as stage IV disease. Two hundred and seventeen patients had received 1st chemotherapy before enrolled in this study. Of these 217 patients, only 6 patients had undergone ctDNA EGFR detection both before and after chemotherapy. Figure 1 showed the scheme of clinical trial design.
EGFR mutation profiles ctDNA samples
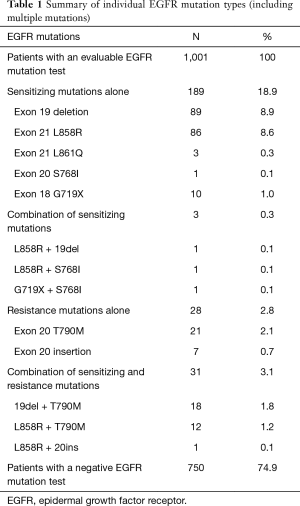
EGFR mutations were detected in 251 of 1,001 (25.1%) patients. The details are summarized in Table 1. Of the 1,001 evaluable samples, 189 (18.9%) harbored sensitizing mutations alone, 28 (2.8%) had resistance mutations [T790M mutation 2.1%, exon 20 insertion (20ins) 0.7%]alone, 3 (0.3%) had a combination of activating mutations, and 31 (3.1%) had a combination of activating and resistance mutations. The most common mutations detected were the exon 19 deletion [deletion alone: 8.9% (89 of 1,001)]; the L858R point mutation in exon 21 [L858R alone: 8.6% (86 of 1,001)]; T790M mutation [2.1% (21 of 1,001)] (Table 1). Of 868 patients with adenocarcinoma, 233 (26.8%) harbored EGFR mutations (86 for ex19del alone, 82 for L858R alone, and 44 for T790M); and of 120 squamous cell lung cancer patients, 14 (11.7%) harbored EGFR mutations (3 for ex19del alone, 2 for L858R alone, and 6 for T790M).
Full table
Influence of chemotherapy on EGFR mutation
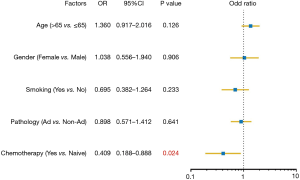
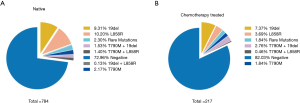
To assess the influence of chemotherapy on ctDNA EGFR mutation, we undertook subgroup analysis. In chemotherapy-naïve patients, EGFR mutation rate was 27.0%, while that was 18.0% in chemotherapy-treated patients (P=0.006). Interestingly, L858R mutation decreased from 10.2% to 3.69% in chemotherapy-treated patients; whereas Exon 19 deletion decreased from 9.31% to 7.37% (Figure 2). Supplementary Table S1 shows the unbalance clinical characteristics between chemotherapy-naive and -treated patients. Since the unbalance existed between chemotherapy-naive and chemotherapy-treated groups, we further undertook multivariate analysis to explore L858R mutation associated factors. We found L858R mutation detection rate significantly decreased after chemotherapy (P=0.024; Figure 3).
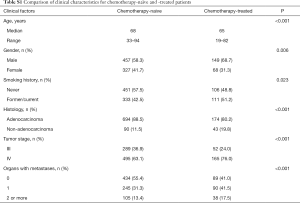
Full table
To further explore the factor correlating with EGFR mutation, multivariate analysis showed gender, histology, chemotherapy, and organ with metastases were independent factors predicting EGFR mutation (Table 2). 22.2% patients without metastasis harbored EGFR mutations, while 24.5% and 37.1% with 1 and ≥2 metastases patients harbored EGFR mutations (P=0.001). Female non-smokers with adenocarcinoma and multiple metastases organs who were chemotherapy-naïve were more prone to carry EGFR mutation. 37.1% of patients with multiple distant metastatic organs detected EGFR mutation, while only 24.5% of patient with thoracic limited metastasis detected EGFR mutation.
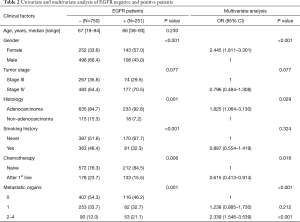
Full table
Treatment choice in clinics after EGFR test
In 254 patients who had the follow-up treatment records, 39 of 56 patients (69.6%) with sensitive EGFR mutations received EGFR-TKI, 114 of 184 (61.9%) patients without sensitive EGFR mutation received chemotherapy ± radiation. Details were in Table 3.
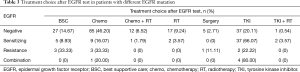
Full table
Discussion
This study is a large-scale real world study of plasma EGFR mutation testing practices in China, which aimed to explore the prevalence of EGFR mutations and determine the correlation of EGFR mutation status with clinical characteristics.
We noticed that the EGFR mutation rate detected in real world practices is generally lower when comparing the data in clinical trials. In China, ARMS method is the only one for ctDNA testing approved by cFDA, the detection sensitive is 1–5% according to the different mutation sites. Although the previous studies showed that the sensitivity ranged from 67.5% to 75% and the specificity is relative high, above 95% by ARMS assay (18). The pooled EGFR mutation detection rate in plasma is about 20% reported in two meta-analysis (19,20). In one real world study, ASSESS, the overall sensitivity is 46%, which is markedly lower than previous reported sensitivity when analyzing the EGFR mutation status between tissue and plasma samples (12). In Japanese population, the EGFR detection rate is 12.0% in 281 plasma samples (12). Our data showed that the overall EGFR mutation rate is 26.8% in 868 adenocarcinoma, and 11.7% in 120 squamous carcinoma in Chinese patients. The relative satisfactory data than ASSESS study revealed the differences in available accessible methodologies, knowledge and quality control, which may contribute to this variation.
In our study, 35 patients had both tissue and plasma samples. The detection concordance was 68.6% (24/35). IGNITE study showed mutation status concordance between 2,581 matched tissue/cytology and plasma samples was 80.5% in overall population and 77.7% in China (21). BENEFIT study showed the concordance was 78.01% with high sensitive droplet digital PCR (22). The relative lower concordance in our studies might be due to the limited number of paired samples and the different time of collection for tissue and plasma.
Currently, more robust platform is available for EGFR mutation detection. The detection sensitivity of the platform can reach 0.1–0.01%. In EGFR mutation analysis of osimertinib phase I registration study, the data showed that sensitivity of sensitive mutation detection is 82–86%, specificity is 96–97% by beaming digital PCR. With droplet digital PCR, the plasma testing sensitivity and specificity were 81.82% and 98.44% for exon19 deletions, 80.00% and 95.77% for L858R. These data indicate the real world mutation analysis in plasma could be further improved if more sensitivity methodology is used.
In this study, we also analyzed the mutation detection rate between naive patients and patient with 1st chemotherapy treatment. This trend is consistent with the data reported in one pooled samples compared with before and after chemotherapy treatment. The lower detection rate was observed in patient treated with chemotherapy. Especially for L858R, the mutation rate decreased obviously compared with other mutation type, from 10.20% to 3.69%, which may indicate that tumor cell with L858R may more sensitive to chemotherapy. Tseng et al. found among patients harboring EGFR mutations, smokers expressed L858R mutation less frequently (35.2% vs. 50.2%, P=0.005) and exon 19 deletions more frequently (52.8% vs. 38.8%, P=0.008) than nonsmokers (23). Although higher percentage of non-smokers was seen in chemotherapy-treated group, our finding showed in non-smoker, L858R mutation also decreased from 14.2% to 4.7% after 1st line chemotherapy. The lux-lung3/6 studies showed that patients with L858R received chemotherapy can obtain longer PFS compared with patients treated with afatinib as 1st line treatment, but not for patients with 19Del (24). Taken together, tumor cell with EGFR L858R may present its distinct sensitivity to chemotherapy and TKI different from 19Del which needs considering more appropriate clinical strategies.
In real-world practice, different stage and metastasis status may significantly complicate the detection rate of EGFR mutation in plasma, considering the release of the ctDNA is influenced by the tumor burden and metastasis. More metastatic tumors generate more DNA leakage into the bloodstream, resulting in higher tumor-derived DNA levels. Previous study showed that higher frequency was found in the plasma of patients with M1b disease than M1a disease (13% vs. 7%) despite the mutation rate is similar in tumor tissue of two groups (25). Mao and his colleagues also found both relative ctDNA abundance and ctDNA concentration had a significant correlation with disease M stage (26). In our study, we further analysis compare the mutation rate among the patients with thoracic limited metastasis, single and multiple distant metastatic organs. Highest mutation rate was observed in patients with multiple distant metastatic organs (37.1%) versus patients with single metastatic organ, patient with thoracic limited metastasis is the lowest (24.5%), which is consistent with the hypothesis of greater release of ctDNA in patients with distant metastases.
Whether ctDNA testing can cause false-positive is always controversy. Tiger X study showed that 23 samples is positive in plasma but negative in tumor tissue, which could be explained by the ctDNA may allow identification of mutations from heterogeneous tumors (27). One recent comprehensive study analyzed the discordance of EGFR mutation status between tumor tissues and matched ctDNA in advanced NSCLC (28). Through ddPCR and NGS further confirmation in biopsy tissues and micro-dissected surgical specimens, 27 negative in tumor, positive in ctNDA patients previous tested were identified harboring EGFR mutation in tissue specimens. These data further indicated that intra and inter-tumor heterogeneity contributed to discordant EGFR mutation status between tissues and ctDNA.
In conclusion, this real-world data suggest that ctDNA is a feasible sample for EGFR mutation analysis when tumor samples are unavailable. It is important to conduct mutation testing with strictly quality control in expert laboratories, using robust and sensitive mutation testing methods to ensure accuracy of results.
Clinical practice points
- The prevalence of EGFR mutations in ctDNA was still unknown in Chinese population.
- Our data from 1001 advanced NSCLC patients showed higher detection rate was observed in chemotherapy-naïve patients than 1st line chemotherapy patients.
- The L858R mutation was significantly lower in patients after 1st line chemotherapy. Tumor cell with L858R mutation may present its distinct sensitivity to chemotherapy.
- Except for gender and histology, number of metastatic organs could influence EGFR mutation.
- This study indicated ctDNA based EGFR mutation test was feasible and could be a surrogate when tissue biopsy is not available.
Acknowledgements
We thank Xun YP and Jiang YP for their work in sample collection and performing EGFR test. We thank Astrazeneca to sponsor this project.
Funding: This study was supported by grants from Projects of Medical and Health Technology in Zhejiang Province (WKJ-2J-1532), the Zhejiang Provincial Natural Science Foundation (LY15H160010) and National Natural Science Foundation of China (Grant no. 81602671).
Footnote
Conflicts of Interest: The abstract of this paper was presented at the IASLC 18th World Conference on Lung Cancer as a poster presentation with interim findings. The poster’s abstract was published in “Poster Abstracts” in Journal of Thoracic Oncology, 12(11):S2212. DOI: https://doi.org/10.1016/j.jtho.2017.09.1472.
Ethical Statement: The protocol was approved by the Institutional Review Board of Hangzhou First People’s Hospital (No. HZFH2015-47-01). All patients signed the informed consent.
References
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Lee CK, Brown C, Gralla RJ, et al. Impact of EGFR inhibitor in non-small cell lung cancer on progression-free and overall survival: a meta-analysis. J Natl Cancer Inst 2013;105:595-605. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Tan DS, Yom SS, Tsao MS, et al. The International Association for the Study of Lung Cancer Consensus Statement on Optimizing Management of EGFR Mutation-Positive Non-Small Cell Lung Cancer: Status in 2016. J Thorac Oncol 2016;11:946-63. [Crossref] [PubMed]
- Fenizia F, De Luca A, Pasquale R, et al. EGFR mutations in lung cancer: from tissue testing to liquid biopsy. Future Oncol 2015;11:1611-23. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Wang S, Han X, Hu X, et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta 2014;430:63-70. [Crossref] [PubMed]
- Jing CW, Wang Z, Cao HX, et al. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev 2014;14:6619-23. [Crossref] [PubMed]
- Zhang H, Liu D, Li S, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn 2013;15:819-26. [Crossref] [PubMed]
- Brevet M, Johnson ML, Azzoli CG, et al. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer 2011;73:96-102. [Crossref] [PubMed]
- Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065-9. [Crossref] [PubMed]
- Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol 2015;36:7-19. [Crossref] [PubMed]
- Wu Y, Liu H, Shi X, et al. Can EGFR mutations in plasma or serum be predictive markers of non-small-cell lung cancer? A meta-analysis. Lung Cancer 2015;88:246-53. [Crossref] [PubMed]
- Mao C, Yuan JQ, Yang ZY, et al. Blood as a Substitute for Tumor Tissue in Detecting EGFR Mutations for Guiding EGFR TKIs Treatment of Nonsmall Cell Lung Cancer: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 2015;94. [Crossref] [PubMed]
- Han B, Tjulandin S, Hagiwara K, et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 2017;113:37-44. [Crossref] [PubMed]
- Wang J, Cheng Y, Wu Y, et al. MA 11.03 Gefitinib as First-Line Treatment of Plasma CtDNA EGFR Mutation-Positive NSCLC Detected by DdPCR: BENEFIT Study (CTONG1405). J Thorac Oncol 2017;12:S1844. [Crossref]
- Tseng JS, Wang CL, Yang TY, et al. Divergent epidermal growth factor receptor mutation patterns between smokers and non-smokers with lung adenocarcinoma. Lung Cancer 2015;90:472-6. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med 2015;372:1700-9. [Crossref] [PubMed]
- Wan R, Wang Z, Lee JJ, et al. Comprehensive Analysis of the Discordance of EGFR Mutation Status between Tumor Tissues and Matched Circulating Tumor DNA in Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1376-87. [Crossref] [PubMed]