Discrepancies between ALK protein disruption and occurrence of ALK gene rearrangement in Polish NSCLC patients
Introduction
Lung cancer (LC) is the most common cause of death among men and women considering cancer mortality in the word. The main causes of LC are smoking and exposure to radon. LC is predominantly detected in an advanced stage with poor prognosis. On molecular level LC is heterogeneous, however some molecular patterns of this disease could be observed. In non-small cell lung cancer (NSCLC), the presence of EGFR, ALK, ROS1, BRAF, MET and HER2 abnormalities might qualify patients to molecularly targeted treatment (in routine practice or in clinical trials).
Epidermal growth factor receptor (EGFR) is a transmembrane protein belonging to the receptor tyrosine kinases family involved in cell survival and proliferation. EGFR gene mutations in tumor cells have been well studied in patients with NSCLC. Approximately 10% of Caucasian patients and 40% of Asian patients showed activating mutations of EGFR gene. Activating mutations occur in exons 18–21, but 90% of them comprise of exon 19 deletions or point mutations in exons 21 (Leu858Arg). EGFR mutations appear mostly in non-smoking, female patients with adenocarcinoma (1). EGFR gene mutations are mutually exclusive with KRAS and ALK genes abnormalities (2). The presence of EGFR mutations is a powerful predictive factor for molecularly targeted treatment with tyrosine kinase inhibitors (TKI) such us erlotinib, gefitinib, afatinib, dacomitinib and osimertinib. Furthermore, some mechanisms of resistance to EGFR-TKIs have been described. Patients with secondary mutation Thr790Met in EGFR gene could be treated with third EGFR-TKIs generation (osimeritinib) (3-6).
Patients without EGFR gene mutations should be diagnosed for rearrangement of anaplastic lymphoma kinase (ALK) gene. Inversion of chromosome 2 [Inv(2)(p21p23)] occurs in approximately 3–5% of patients with NSCLC, more often in young, non-smoking, male patients with adenocarcinoma (median age 52 years). For ALK gene, the most common partner for fusion is EML4 gene (echinoderm microtubule associated protein like 4). Protein product of fusion genes is constitutively active, which leads to increased cells proliferation, growth and survival. In NSCLC other partners for ALK rearrangement have been described: TFG, KIF5B (Kinesin Family Member 5B), DCTN1 (Dynactin Subunit 1) and SQSTM1 (Sequestosome 1) genes. Presence of ALK gene rearrangement is an indication for targeted therapy based on ALK inhibitors (crizotinib, ceritinib or alectinib) (2,7-11).
Despite many years of experience in analysis of these genetic abnormalities, there are still problems in interpretation of molecular tests results (12). DNA degradation during the thermal and chemical processing of tumor materials and scarcity of material may unable the performance of molecular tests for EGFR gene mutations (non-diagnostic or false negative results of real-time PCR analysis) or for ALK gene rearrangement (non-diagnostic results of fluorescent in situ hybridisation, FISH). In contrast, different ALK gene abnormalities in a small tumor cell clone may affect the expression of abnormal ALK protein on the small population of tumor cells, which is visualised in immunohistochemistry (IHC) tests. This causes false positive results of IHC tests (confirmation of ALK gene rearrangement involves presence of 15% of nuclei with ALK abnormalities detected in FISH method)
The aim of this study was the identification of the most important diagnostic problems in the routine analysis of ALK abnormalities in NSCLC patients. We assessed the occurrence of EGFR and ALK disruption in large group of Caucasian NSCLC patients. The analysis of relationship between genetic abnormalities presence and patients’ demographic and clinical features was also performed.
Methods
Patients
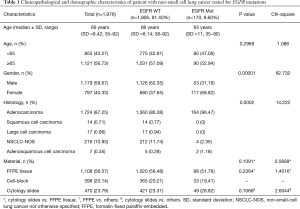
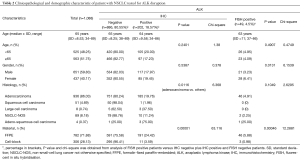
A total of 1,976 NSCLC patients were tested for EGFR mutations using real-time PCR method form February 2016 to June 2017. DNA was isolated from 1,108 formalin-fixed paraffin-embedded (FFPE) tumor tissues and 868 cytological specimens (398 cell-blocks and 470 cytological slides). After excluding the presence of EGFR gene mutation, 1,100 NSCLC patients were examined for ALK abnormalities. Patients with wild-type (WT) EGFR gene and with sufficient tissue or cellular materials archived in FFPE blocks were qualified to diagnosis of ALK abnormalities using IHC test. IHC examination was performed in 782 histological and 306 cytological (cell-blocks) samples. ALK gene rearrangement using FISH method was analysed only in IHC positive cases. Demographic and clinic-pathological characteristics of the patients were compared with the score of molecular and immunohistochemical analysis (Tables 1,2).
Full table
Full table
Any aspect of the work covered in this manuscript was approved by the Ethics Committee of the Medical University of Lublin, Poland (No. KE-0254/169/2014).
Real-time PCR analysis of EGFR gene mutations
EGFR gene mutations analysis was carried out when the presence of more than 10% of tumour cells was observed by a pathologist in H & E slides. DNA was extracted using QIAamp DNA FFPE Tissue Kit (CE-IVD marked, Qiagen, Germany). Isolation was performed according to the manufacturer’s instructions. Concentration and quality of isolated DNA was estimated by spectrophotometry.
Mutations of EGFR gene (NM_005228.3) were identified using the EntroGen EGFR mutations Analysis Kit (USA) on Cobas Z480 real-time PCR system (Roche Diagnostics, USA). The following mutations were examined (in exons 18 to 21): p.Glu709Asp (c.2127A>C), p.Glu709Ala (c.2126A>C), p.Glu709Gly (c.2126A>G), p.Glu709Lys (c.2125G>A), p.Glu709Gln (c.2125G>C), p.Glu709Val (c.2126A>T), p.Gly719Ala (c.2156G>C), p.Gly719Ser (c.2155G>A), p.Gly719Cys (c.2155G>T); c.2235-2249 del 15, c.2235-2252>AAT del 18, c.2236-2253 del 18, c.2237-2251 del 15, c.2237-2254 del 18, c.2237-2255>T del 19, c.2236-2250 del 15, c.2238-2255 del 18, c.2238-2248>GC del 11, c.2238-2252>GCA del 15, c.2233-2247 del 15, c.2234-2248 del 15, c.2235-2246 del 12, c.2235-2248>AATTC, c.2235-2251>AATTC, c.2235-2252>AAT, c.2235-2255>AAT, c.2236-2248>AGAC, c.2236-2248>CAAC, c.2236-2256 del 21, c.2237-2252>T, c.2239-2247 del 9, c.2239-2256 del 18, c.2239-2248>C del 10, c.2239-2258>CA del 20, c.2240-2251 del 12, c.2240-2257 del 18, c.2240-2254 del 15, c.2239-2251>C del 13, c.2237-2253>TC, c.2237-2253>TTCCT, c.2237-2253>TTGCT, c.2237-2256>TC, c.2237-2256>TT, c.2237-2257>TCT, c.2238-2252 del 15, c.2239-2252>CA, c.2239-2253 del 15, c.2239-2256>CAA, c.2239-2257>T, c.2239-2262 del 24, c.2246-2260 del 15, c.2248-2273>CC, c.2252-2275 del 24, c.2252-2276>A, c.2252-2277>AT, c.2253-2276 del 24, c.2254-2277 del 24, p.Thr790Met (c.2369C>T), p.Ser768Ile (c.2303G>T), c.2307-2308 ins GCCAGCGTG, c.2319-2320 ins CAC, c.2310-2311 ins GGT; p.Leu858Arg (c.2573T>G), p.Leu858Met (c.2572C>A), p.Leu861Gln (c.2582T>A), p.Leu861Arg (c.2582T>G). The analysis of EGFR mutations has been performed simultaneously with the positive and negative control examination, according to the manufacturer protocol.
IHC test of abnormal ALK protein expression
Abnormal ALK protein expression was examined using IHC test. Analyses were performed on 3 µm sections of paraffin-embedded tissue, fixed on Thermo Scientific Superfrost Plus™ glass slides. Tissue sections on glass slides were preheated in 56 °C on hotplate for at least 3 hours.
ALK protein IHC staining was conducted on Ventana Benchmark GX platform, using CE-IVD approved anti-ALK Rabbit Monoclonal Primary Antibody (clone D5F3). OptiView Amplification Kit and OptiView DAB IHC Detection Kit were used as a detection system. Hematoxylin counterstaining was incorporated in the staining protocol. As a negative control Rabbit monoclonal negative control immunoglobulin was used (Ventana Medical System, Tuscon, USA) (12).
After staining all glass slides were washed and then dehydrated in a series of two 96% ethanol and two xylene washing steps. The last step was coverslipping of the glass slides. The staining analysis was performed according to the International Association for the Study of Lung Cancer (IASLC) and Ventana (Roche, USA) guidelines (12).
FISH analysis of ALK gene rearrangement
All positive results obtained in IHC staining were re-evaluated by FISH method to visualize the presence of ALK rearrangement using the Vysis ALK Break Apart FISH Probe Kit (Abbot Molecular, USA), paraffin-pretreatment IV and Post-Hybridization Wash Buffer Kit (Abbot Molecular, USA) in fluorescent microscope (Nikon Eclipse 55i, Japan). The localization and content of tumor cells were examined with H & E staining in serially prepared slides. Interpretation of FISH results was conducted in accordance to American Food and Drug Administration (FDA) and IASLC guidelines (13).
Statistical analysis
Statistical analysis was performed using Statistica v.12. Associations between presence of EGFR mutations, abnormal ALK protein expression, ALK gene rearrangement and clinical factors as well as the type of tumor materials were examined using Pearson’s chi-square test. The student-t, U-Mann-Whitey and Kruskal-Wallis ANOVA tests were used for testing equality of population means and medians among subgroups. Spearman correlation was used for assessment of the relationship between IHC and FISH results. P values below 0.05 were considered significant.
Results
EGFR mutation analysis
In group of 1,976 NSCLC Caucasian patients, 170 (8.6%) had mutations in EGFR gene, including 164 adenocarcinoma patients (9.51% of all adenocarcinoma patients). Characteristic of patients with available results of EGFR gene analysis is showed in Table 1.
Obviously, EGFR gene mutations were significantly more common in women than in men and in adenocarcinoma than in other types of NSCLC. Surprisingly, mutations in EGFR gene were slightly more common in cytological slides containing few cancer cells than in the FFPE tissue and cell-blocks (chemical and thermal processing of cytological slides is not that harmful than FFPE materials) (Table 1).
Deletions in exon 19 and Leu858Arg substitution in exon 21 of EGFR gene were diagnosed in 84 and 65 cases, respectively (49% and 38% of all tested EGFR mutations, respectively). Rare EGFR mutations were: substitution Gly719X in 10 cases (5.9%), substitution Leu861Gln in 7 cases (4.1%), insertions in exon 20 in 6 cases (3.5%), substitution Glu709X in 5 cases (2.9%), primary substitution Thr790Met in 5 cases (2.9%) and substitution Ser768Ile in 3 cases (1.8%). In 8 cases (4.7%), coexistence of two different EGFR gene mutations were detected (in two cases Gly719X and Ser761Ile substitutions, in two cases Gly719X and primary Thr790Met substitutions, in two cases deletions in exon 19 and primary Thr790Met substitution, and in individual cases—Leu858Arg and primary Thr790Met substitutions as well as Leu858Arg and Ser768Ile substitutions). Using covariance analysis of female patients with EGFR gene mutations, we showed that patients with Leu858Arg and Leu861Gln substitutions as well as exon 20 insertions were significantly older than patients with deletions in exon 19, Glu709X and Ser761Ile substitutions. Moreover, female patients with deletions in exon 19 of EGFR gene were significantly younger than women without EGFR gene mutations (P=0.0054). On the other hand, female patients with Leu858Arg substitution were significantly older than female patients without EGFR gene mutations (P=0.000093). Similar differences have not been demonstrated in male NSCLC population.
Analysis of abnormal ALK protein expression and ALK gene rearrangement
Analysis of ALK disruption was possible in 1,100 NSCLC patients of 1,806 cases tested for EGFR gene mutations and with wild type of EGFR gene (60.9%). Using IHC test, we detected abnormal ALK protein expression in 202 patients (18.57% of patients with ALK IHC test). IHC test was non-diagnostic in 12 patients (1.1% of patients studied cases). Abnormal ALK protein expression was observed with similar frequency in men and in women, as well as in older and younger patients. Adenocarcinoma patients expressed abnormal ALK protein significantly more often than other NSCLC patients. However, expression of abnormal ALK protein was also observed in particular patients with adeno-squamous cell carcinoma and large cell carcinoma. ALK expression was detected significantly more often in FFPE tissue than in cell-blocks (Table 2).
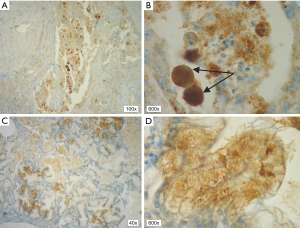
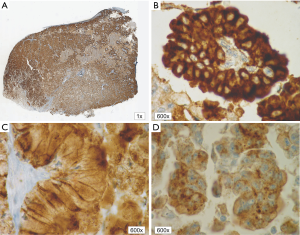
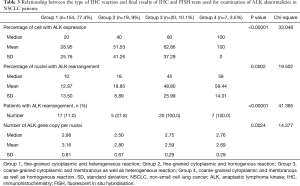
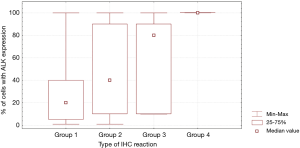
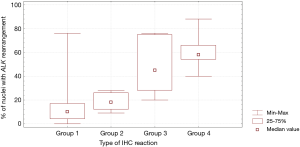
In patients with ALK expression, data about percentage of neoplastic cells with abnormal ALK protein expression and type of IHC reaction were available in 199 cases. Median percentage of neoplastic cells expressing disrupted ALK was 30%±34.62% in the whole studied group, 40%±38% in female patients, and 25%±31% in male patients. Expression of abnormal ALK protein on less than 15% of tumor cells was observed in 73 patients of 199 cases (36.7%). One hundred and seventy-two patients (86%) had fine-grained cytoplasmic type of IHC reaction (cytoplasmic stippling), while 27 patients (14%) had coarse-grained cytoplasmic and membranous type of IHC reaction. Homogeneous (same type and intensity of IHC reaction in part of tumor where ALK expression was found) and heterogeneous character of IHC reaction was observed in 25 (12.6%) and 174 (87.4%) patients with positive result of IHC test. Patients with cytoplasmic stippling and heterogeneous character of IHC reaction were significantly older than patients with coarse-grained cytoplasmic and membranous as well as homogenous type of IHC reaction (P<0.005). Detailed characteristic of type of IHC reaction revealed: fine-grained cytoplasmic (cytoplasmic stippling) and heterogeneous reaction in 154 patients (77.4%, group 1), fine-grained cytoplasmic (cytoplasmic stippling) and homogenous reaction in 18 patients (9%, group 2), coarse-grained cytoplasmic and membranous as well as heterogeneous reaction in 20 patients (10.1%, group 3), coarse-grained cytoplasmic and membranous as well as homogenous reaction in 7 patients (3.5%, group 4). An example of abnormal ALK protein expression and non-specific staining in IHC test were presented in Figures 1-4. Percentage of cells with abnormal ALK protein expression was significantly (P<0.00001) lower in group 1 and 2 compared to group 3 and 4 (Table 3, Figure 5).
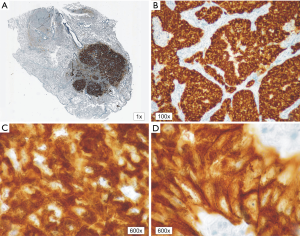
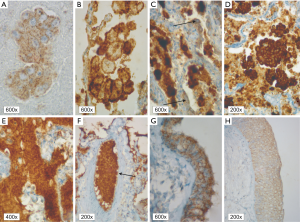
Full table
Using FISH technique, we confirmed the presence of ALK gene rearrangement in 49 patients (4.5% of the whole group of patients and 24.26% of patients with expression of abnormal ALK protein detected in IHC method). Thus, the positive predictive value of the IHC technique in our group of NSCLC patients was only 24.26%. The FISH results were non-diagnostic in 21 patients (11.5% of ALK IHC positive patients). ALK rearrangement was detected more often in women than in men and in FFPE tissue than in cell-blocks. Characteristic of ALK rearranged patient is summarized in Table 2.
In the whole FISH group, the median percentage of nuclei with ALK rearrangement was 12%±19.17%. It should be noted that only 18 patients (9.8% of patients with valid result of FISH test) showed no tumor nuclei without ALK gene rearrangement. In patients with diagnosis of ALK gene rearrangement and qualified to ALK inhibitors therapy (≥15% of nuclei with ALK gene rearrangement), the median percentage of nuclei with ALK rearrangement was only 25.5%±19.72%. ALK rearrangement defined as coexistence of single red and split signals was observed in 27 patients (55.1% of patients with FISH positive results), only split signals were observed in 1 case (2% of patients with FISH positive results) and only single red signals were diagnosed in 21 patients (42.9% of patients with FISH positive results). The polysomy (≥4 gene copy number per nuclei) of ALK gene was observed in 39 patients (21.4% of patients with diagnostic results of FISH examination). Median number of ALK gene copy per nuclei was 2.9±0.77.
Type and intensity of IHC reaction used for detection of abnormal ALK protein affected the results of the FISH analysis. All 27 patients with coarse-grained cytoplasmic and membranous as well as homogenous or heterogenous type of IHC reaction had positive result of FISH analysis (100%). While, patients with fine-grained cytoplasmic (cytoplasmic stippling) type of IHC reaction (especially with heterogenous type of IHC staining) rarely demonstrated ALK gene rearrangement in more than 15% of tumor nuclei (P<0.00001, Table 3). Moreover, patients from group 3 and 4 had significantly higher percentage of nuclei with ALK gene rearrangement than patients from group 1 and 2 (P=0.0002, Table 3, Figure 6). All 21 patients without any nucleus with ALK gene rearrangement had fine-grained cytoplasmic and heterogeneous character of IHC reaction. It should be assumed that the IHC results in these patients were completely false. There was significant positive correlation between percentage of tumor cells with expression of ALK abnormal protein detected in IHC test and percentage of nuclei with ALK gene rearrangement visualised in FISH analysis (R =+0.617, P<0.00001).
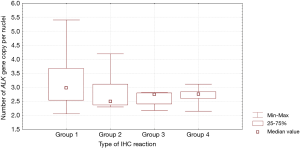
Patients with fine-grained cytoplasmic (cytoplasmic stippling) and heterogenous type of IHC reaction showed highest number of ALK gene copy per nuclei analysed with FISH method. Patients from group 1 and 2 had significantly higher number of ALK gene copy than patients from group 3 and 4 (P=0.0024, Table 3, Figure 7). Moreover, we found significant negative correlation between percentage of tumor cells with expression of ALK abnormal protein detected in IHC test and number of ALK gene copy per nuclei visualised in FISH analysis (R =–0,2004; P<0.05). It should be noted that in the FISH method, patients with ALK gene rearrangement most often showed normal ALK gene copy number (median 2.82 ALK gene copy per nuclei).
Discussion
Molecular changes in NSCLC could be observed on epigenetic, genetic and protein level. Nowadays, detection of EGFR gene mutations and ALK disruption is substantial for implementation of molecularly targeted treatment. Other methods of treatment (immunotherapy and chemotherapy) in locally advanced or advanced NSCLC patients with EGFR gene mutations or ALK gene rearrangement can be considered only after exhaustion of TKI EGFR or ALK treatment possibilities (13).
The frequency of EGFR gene mutations in NSCLC patients is variable, depending of the population. Werutsky et al. indicated that the median global prevalence of EGFR mutations in NSCLC patients is 33.07% (14). In Europe, Midha et al. pointed that overall frequency of EGFR mutations in adenocarcinoma patients is 15% (range, 6–41%) (1). However, Boch et al. indicated a lower frequency (4.9%) for EGFR mutations in an unselected Caucasian patients with NSCLC (including squamous cell carcinoma) and noticed that clinical trials may overestimate EGFR mutations frequency in NSCLC due to important selection biases (15). In our study group, we detected EGFR mutations in 8.6% of patients qualified to EGFR-TKIs treatment (9.51% of adenocarcinoma patients showed EGFR gene mutations). Therefore, the prevalence of EGFR mutations in the Polish population is similar to the frequency of this genetic abnormality in analogous populations from Sweden (10% of NSCLC patients, 11.5% of adenocarcinoma patients), Germany (8.7% of adenocarcinoma patients), Denmark (8% of adenocarcinoma patients) or Italy (9% of NSCLC patients, 12.6% of adenocarcinoma patients) (15-18). In most of these populations, EGFR gene mutations are more common in non-smokers and in female patients as well as could be detected in any material containing the appropriate number and percentage of cancer cells (FFPE material or cytology slides) (1,14-19). However, cytology slides contain the best DNA quality and may be an even better material for EGFR mutations analysis if the errors in the preparation of FFPE tissue or cell-blocks occur.
In this article and in our previous studies, we reported that rare EGFR mutations are not so unique. Similarly to other populations of NSCLC patients, in our population, the deletions in exon 19 (49%) and Leu858Arg substitution in exon 21 (38%) were the most common, but rare EGFR mutations occurred in 13% of patients (20). Globally, Werutsky et al. indicated that 54.5% patients with EGFR gene mutations showed exon 19 deletions, 36.4% —exon 21 substitution (Leu858Arg) and 9.1%—rare mutations (14). Our original observation is the relationship between the occurrence of different EGFR mutations and the age of female patients with NSCLC. Older women were more predisposed to Leu858Arg and Leu851Gln substitutions or insertions in exon 20, younger—to exon 19 deletions.
In diagnostic algorithm used in qualification of NSCLC patients to molecularly targeted therapies, the next step after EGFR mutations analysis is identification of ALK disruption. Approximately 5.7% of adenocarcinoma patients showed ALK gene rearrangement. Among more than 12,000 NSCLC patients described in literature, ALK gene rearrangement was found mainly in non-squamous and non-endocrine LC patients, as well as in never smokers or light ex-smokers, young (40–50 years old), male patients (12,21-23). In our study, we identified ALK rearrangement in 4.5% of NSCLC patients (4.91% of adenocarcinoma patients showed ALK rearrangement) qualified routinely to ALK inhibitors therapy. Surprisingly, our population of patients with ALK rearrangement was similar in age (63 years old) to patients with EGFR mutation (65 years old) and to overall population (66 years old). Moreover, ALK gene rearrangement was more common in women.
The proper diagnosis of ALK abnormalities is most important for the correct qualification for therapy with ALK inhibitors in NSCLC patients. Interpretation of IHC and FISH results yields much more problems than the interpretation of real-time PCR results. The diagnosis of ALK abnormalities is characterized by the subjectivism of the microscopic slides observers, which is eliminated in EGFR gene mutations diagnosis.
IHC holds promise as a reliable screening method for ALK disruption before confirmation of ALK gene rearrangement with FISH technique (24). Specificity and sensitivity of IHC method with D5F3 monoclonal antibody clone (also used in this study) has been compared with FISH method in FFPE histological material. The sensitivity ranged from 81% to 100%, and the specificity, form 82% to 100% (12,21,24-28). Wang et al. found that IHC test with D5F3 clone used for ALK expression diagnosis on cell-blocks from pleural effusion was 100% concordant with FISH results of ALK gene rearrangement (29). Other authors showed that IHC tests in cell-blocks from fine-needle biopsy materials were associated with good specificity but low sensitivity (12). Our results indicate that ALK gene rearrangement could not be confirmed in a large percentage of materials with abnormal ALK protein expression (false-negative results and low specificity of IHC test). On the other hand, we found low percentage of cell-blocks with expression of abnormal ALK protein, which may indicate low sensitivity of the IHC test in the fine-needle biopsy material.
The Pathology Committee of the IASLC published guidelines, which can help pathologists, diagnosticians and oncologists to better understand the background, protocols and interpretation of ALK testing results in NSCLC patients (12). However, some recommendations for ALK IHC diagnosis are not precise. Tumor cells with ALK rearrangement usually express fusion protein consisting of ALK and EML4 proteins. Expression of this fusion protein is cytoplasmic. IHC staining has a granular character and in some cases membranous reaction is also observed. The assessment of staining intensity is subjective. Some authors have scored the intensity from 1+ to 3+ with an ambiguous threshold around 1+ or 2+. Other pathologists have defined positive ALK expression as more than 10% of tumor cells with any expression of ALK fusion protein, regardless of the IHC reaction intensity (12,21,24-28,30). This confirms that some tumors are heterogeneous regarding ALK or other genes expression. However, Takeuchi et al. found that almost all cancer cells expressed ALK fusion protein (homogenous IHC reaction), if ALK gene rearrangement was found in tumor cells nuclei. They concluded that the heterogeneity is likely related to heterogeneity of the material fixation (31). The results of other authors, including our results, confirmed observation of significant heterogeneity of ALK IHC staining. Moreover, ALK gene rearrangement was observed in single cell nuclei in many patients, which confirmed the heterogeneity of tumors also at the molecular level. Camidge et al. found that in approximately 8% of NSCLC patients, the rate of rearrangement-positive nuclei ranged within 10% to 20% (32). Therefore, the percentage of 15% of nuclei with ALK gene rearrangement was arbitrarily assumed as a positive FISH test result (12,21,24-28).
There are some artifacts that may lead to false-positive ALK IHC staining: light cytoplasmic stippling, positive IHC reaction in alveolar macrophages and in cells of neural origin (including neuroendocrine cells) as well as glandular epithelia, extracellular mucin and necrotic areas staining (12,21,24-28). Some researchers showed that membranous staining in ALK IHC test was more frequent in patients without ALK rearrangement (24,33). Moreover, ALK protein expression may be increased in some situations without ALK gene rearrangement. Salido et al. discussed association between ALK gene copy number and ALK protein expression and its clinical implications. They found ALK gene amplification in 63% of adenocarcinoma patients and 17% of squamous cell carcinoma patients (34).
Our results are moderately consistent with the cited observations. In our study, membranous staining in ALK IHC test was associated with ALK gene rearrangement. Cells with light cytoplasmic stippling in IHC staining were predominantly negative for ALK rearrangement in FISH analysis. However, even in this group of patients, FISH-positive-patients were diagnosed. On the other hand, the percentage of nuclei with ALK gene rearrangement in this group of patients was low (about 15%) and the patients may not benefit from the therapy with ALK inhibitors. We have found that cytoplasmic stippling may be associated with ALK gene amplification. In addition, we confirmed that gain of ALK gene copy number is very common in adenocarcinoma patients. We showed in our study 202 cases positive in IHC and 49 cases in FISH which is 76% discordance between these methods of detection. Alì et al. indicated in their study that among 523 tumor samples analyzed 20 (3.8%) were positive for ALK rearrangement in FISH analysis, 18 (3.4%) were ALK positive in IHC, 18 samples (3.4%) had concordant IHC and FISH results. In 2 (0.3%) of ALK positive cases in FISH method they did not observe ALK protein expression in IHC. In this 2 discrepant cases, they did not detect any mass peaks for the EML4-ALK variants in MassARRAY (35).
Furthermore, Cabillic et al. demonstrated in large group of 3,244 NSCLC patients, at two independent French centers, FISH-positive and/or IHC-positive results in 150 (4.6%) cases. Only 80 of 150 specimens were ALK positive in both techniques. Thirty-six cases were FISH-positive/IHC-negative, 19 were FISH-negative/IHC-positive and 15 were FISH-non-contributive/IHC-positive (36). Cabillic et al. indicated that a single FISH or IHC analysis conducted merely would have failed to detect approximately one-fourth of the ALK-positive cases. They concluded that there is a necessity of combined testing of ALK disorders in anti-ALK TKI treatment qualification. Yatabe et al. indicating that discrepancies in IHC and FISH method can be caused by nonspecific staining or high-grade neuroendocrine tumor (IHC false positive results), mucin-rich cells (signet ring cells) or technical issues for example poor fixation, insensitive detection method (IHC false negative), thick paraffin sections which can cause separation of FISH signals on the sectioning surface, resulting in imitated break-apart signals (FISH positive results), and compressed z-stacked images, atypical signal profile (FISH false negative results) (37). In our study discrepancies between FISH and IHC method were higher than described by Alì et al. or Cabillic et al. (35,36). Yatabe et al. pointed out several causes that might affect the discrepancies in the results and we are in line with them (37). However, we concluded that there is a necessity for careful consideration of the antibody used in ALK protein detection in the IHC method. Maybe, it should be more specific for abnormal ALK protein detection. Discrepancies between the two methods may occur in certain cases because of disturbance in abnormal ALK protein expression. However, after the end of our study, the producer of IHC kit for the detection of abnormal ALK protein expression halted the sale of reagents. False positive IHC results have appeared in many European laboratories.
It is interesting that Cabillic et al. and Alì et al. shoved ALK gene rearrangement but they did not detect expression of abnormal ALK protein (35,36). It indicated that other genetic mechanisms (other than EML4 gene partners for ALK translocation) could have influenced ALK rearrangement occurrence without simultaneous abnormal ALK protein expression. In our investigation we did not observe cases positive in FISH method and negative in IHC test. In routine diagnostics, we first diagnose the expression of an abnormal ALK protein, and in case of a positive result in the IHC test, we confirm the presence of ALK gene rearrangements with FISH method.
We observed a large percentage of non-diagnostic FISH results (lack of probe hybridization, high background noise and stringy signals, as well as limited percentage of nuclei with ALK rearrangement). In such cases, it would be recommended to use other diagnostic methods for examination of ALK gene abnormalities. In Poland, reimbursement of ALK inhibitor is limited to patients with ALK rearrangement detected with FISH technique, therefore other techniques are rarely used and FISH method remains a gold standard in laboratory practice for ALK gene rearrangement detection. However, in the near future, we should expect wider use of RT-PCR (reverse transcription-polymerase chain reaction) and next generation sequencing (NGS) techniques in ALK abnormalities diagnosis.
Conclusions
EGFR gene mutations occurrence in our NSCLC patients group was similar to other European populations. However, Polish patients were characterized by a high prevalence of rare EGFR gene mutations. We indicated that diagnosis of ALK disruption in NSCLC patients should be notably careful using IHC and FISH methods. Difficulties in the interpretation of the results of these tests correlated with high percentage of patients expressing abnormal ALK protein or artifacts in the IHC method and a high percentage of patients with non-diagnostic results of the ALK rearrangement examination with FISH method. The frequency of ALK rearrangement in our population was similar to other Caucasian groups of NSCLC patients. However, ALK gene rearrangement was observed in older population than in other studies. We concluded that the recommendations for ALK diagnosis should include the way of interpretation of cases with low percentage of abnormal ALK protein-expressing cells in IHC test, with different character of IHC reaction, with low percentage of ALK gene rearranged nuclei, and gain of ALK gene copy number in FISH method.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Any aspect of the work covered in this manuscript was approved by the Ethics Committee of the Medical University of Lublin, Poland (No. KE-0254/169/2014).
References
- Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PubMed]
- Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 2010;17:889-97. [Crossref] [PubMed]
- Haaland B, Tan PS, de Castro G Jr, et al. Meta-analysis of first-line therapies in advanced non-small-cell lung cancer harboring EGFR-activating mutations. J Thorac Oncol 2014;9:805-11. [Crossref] [PubMed]
- Wu YL, Cheng Y, Zhou X, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454-66. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Mok TS, Wu Y-L, Ahn M-J, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. Erratum in: N Engl J Med 2015;373:1582. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. Erratum in: N Engl J Med 2015;373:1582. [Crossref] [PubMed]
- Soria JC, Tan DSW, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 2017;389:917-29. Erratum in: Lancet 2017;389:908. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Tsao MS, Hirsch FR, Yatabe Y. IASLC Atlas of ALK and ROS1 testing in lung cancer. (Second Edition). Editorial Rx Press, 2016.
- Blackhall F, Cappuzzo F. Crizotinib: from discovery to accelerated development to front-line treatment. Ann Oncol 2016;27 Suppl 3:iii35-41. [Crossref] [PubMed]
- Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-v27. [Crossref] [PubMed]
- Werutsky G, Debiasi M, Sampaio FH, et al. P1.08: Updated Analysis of Global Epidemiology of EGFR Mutation in Advanced Non-Small Cell Lung Cancer: Track: Prevention, Early Detection, Epidemiology and Tobacco Control. J Thorac Oncol 2016;11:S184-5. [Crossref] [PubMed]
- Boch C, Kollmeier J, Roth A, et al. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC): routine screening data for central Europe from a cohort study. BMJ Open 2013;3. [Crossref] [PubMed]
- Sandelin M, Berglund A, Sundström M, et al. Patients with Non-small Cell Lung Cancer Analyzed for EGFR: Adherence to Guidelines, Prevalence and Outcome. Anticancer Res 2015;35:3979-85. [PubMed]
- Pasello G, Polo V, Frega S, et al. 166P: Non-small cell lung cancer (NSCLC) patients with rare or complex epidermal growth factor receptor (EGFR) mutations: A single institution series. J Thorac Oncol 2016;11:S130. [Crossref]
- Skov BG, Høgdall E, Clementsen P, et al. The prevalence of EGFR mutations in non-small cell lung cancer in an unselected Caucasian population. APMIS 2015;123:108-15. [Crossref] [PubMed]
- Boldrini L, Alì G, Gisfredi S, et al. Epidermal growth factor receptor and K-RAS mutations in 411 lung adenocarcinoma: a population-based prospective study. Oncol Rep 2009;22:683-91. [PubMed]
- Krawczyk P, Reszka K, Ramlau R, et al. Prevalence of rare EGFR gene mutations in nonsmall-cell lung cancer: a multicenter study on 3856 Polish Caucasian patients. Ann Oncol 2016;27:358-9. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823-59. Erratum in: J Thorac Oncol 2013;8:1343. [Crossref] [PubMed]
- An SJ, Chen ZH, Su J, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One 2012;7. [Crossref] [PubMed]
- Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 2009;15:5216-23. Erratum in: Clin Cancer Res 2009;15:7110. [Crossref] [PubMed]
- Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 2010;16:1561-71. [Crossref] [PubMed]
- Conklin CM, Craddock KJ, Have C, et al. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013;8:45-51. [Crossref] [PubMed]
- McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol 2012;7:348-54. [Crossref] [PubMed]
- Martinez P, Hernández-Losa J, Montero MÁ, et al. Fluorescence in situ hybridization and immunohistochemistry as diagnostic methods for ALK positive non-small cell lung cancer patients. PLoS One 2013;8. [Crossref] [PubMed]
- Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:322-8. [Crossref] [PubMed]
- Wang W, Tang Y, Li J, et al. Detection of ALK rearrangements in malignant pleural effusion cell blocks from patients with advanced non-small cell lung cancer: a comparison of Ventana immunohistochemistry and fluorescence in situ hybridization. Cancer Cytopathol 2015;123:117-22. [Crossref] [PubMed]
- Jiang L, Yang H, He P, et al. Improving Selection Criteria for ALK Inhibitor Therapy in Non-Small Cell Lung Cancer: A Pooled-Data Analysis on Diagnostic Operating Characteristics of Immunohistochemistry. Am J Surg Pathol 2016;40:697-703. [Crossref] [PubMed]
- Takeuchi K. Interpretation of anti-ALK immunohistochemistry results. J Thorac Oncol 2013;8:e67-8. [Crossref] [PubMed]
- Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-9. [Crossref] [PubMed]
- Murakami Y, Mitsudomi T, Yatabe Y. A Screening Method for the ALK Fusion Gene in NSCLC. Front Oncol 2012;2:24. [Crossref] [PubMed]
- Salido M, Pijuan L, Martínez-Avilés L, et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol. 2011;6:21-7. [Crossref] [PubMed]
- Alì G, Proietti A, Pelliccioni S, et al. ALK rearrangement in a large series of consecutive non-small cell lung cancers: comparison between a new immunohistochemical approach and fluorescence in situ hybridization for the screening of patients eligible for crizotinib treatment. Arch Pathol Lab Med 2014;138:1449-58. [Crossref] [PubMed]
- Cabillic F, Gros A, Dugay F, et al. Parallel FISH and immunohistochemical studies of ALK status in 3244 non-small-cell lung cancers reveal major discordances. J Thorac Oncol 2014;9:295-306. [Crossref] [PubMed]
- Yatabe Y. ALK FISH and IHC: you cannot have one without the other. J Thorac Oncol 2015;10:548-50. [Crossref] [PubMed]


