Comparison of MALDI-TOF MS, gene sequencing and the Vitek 2 for identification of seventy-three clinical isolates of enteropathogens
Introduction
Bacterial diarrhea is induced by a number of different pathogens, including Salmonella, Escherichia coli, Shigelle, Campylobater, Aeromonas, Plesiomonas shigelloides, Yersinia enterocolitica, and pathogenic Vibrio, etc. The microbiologic etiology of diarrhea is often not clinically obvious, thus, the rapid and sensitive identification of the enteropathogens in the clinical laboratory is essential for early and accurate diagnosis and timely therapy. For decades, enteropathogens have been routinely identified in clinical microbiology laboratories with biochemical methods and serological tests which are usually time-consuming and require large amounts of biological materials. Moreover, species identification based on biochemical test results is sometime unreliable and gives controversial results. Molecular methods have been demonstrated to have complementary value, however, the high cost and high technical expertise required have hindered their routine uses in clinical laboratories. More recently, however, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been developed for identification of microorganisms based on the comparison of sample mass spectral fingerprints that are unique protein signatures for each microorganism to reference mass spectral fingerprints. One or a set of characteristic peaks in the mass spectral fingerprints, which appear to be conserved for a certain type of microorganism, are used for the identification. Since being commercialized in 2008, MALDI-TOF MS has been evaluated in terms of its analytical accuracy to identify clinical isolates by many diagnostic laboratories.
A couple MALDI-TOF MS systems have entered the market of microbial identification tools. It was reported that MALDI-TOF MS provided rapid and reliable identifications of medical bacteria and fungi isolated from patients (1). However, the analytical accuracy varies according to isolate species and the different MS systems, and it was suggested that unsatisfactory identifications of some strains were related mainly to the databases or known limitations of the MALDI-TOF MS technique (2). The application of MALDI-TOF MS in identification of routine isolates in clinical laboratories has been reported (3,4), but its performance for enteropathogens is not as clear. We report here the clinical evaluation of MALDI-TOF MS for identification of enteropathogens.
Methods
Bacteria and culture conditions
Salmonella enterica (ATCC14028), Shigelle sonnei (GIM1.239), S. flexneri (GIM1.238), S. dysenteriae (GIM1.236), Campylobacter jejuni (ATCC33291), Aeromonas hydrophila (GIM1.172), Yersinia enterocolitica (GIM1.265), Vibrio parahaemolyticus (ATCC17802), V. fluvialis (GIM1.488) were purchased from the Guangdong Provincial Institute of Microbial Culture Collection (China), and the V. cholera attenuated strain was provided by Guangzhou CDC (China). Quality control strain of Escherichia coli (ATCC8739) for the Vitek MS system was provided by bioMérieux (France). Seventy-three clinical isolates of enteropathogens covering seven different genera were isolated from patients with diarrheal disease between October 2012 and September 2013 in our hospital. Bacteria were recovered from –80 °C. Clostridium difficile was cultured anaerobically on anaerobic blood agar at 35 °C for 48 h. All others were cultured on Columbia blood agar but under different conditions: Salmonella, Shigelle, Aeromonas, P. shigelloides, V. parahaemolyticus, V. fluvialis, and the V. cholera attenuated strain were cultured at 35 °C in 5% CO2 for 24 h; Campylobacter strains at 42 °C in microaerophilic condition for 48 h; and Y. enterocolitica at 28 °C for 48 h. All culture media were purchased from Guangzhou Detgerm Microbiology Technology (China).
MALDI-TOF MS analysis
A portion of a fresh colony of the bacterium was smeared onto a 48-well target plate and then immediately covered with 1 µL of α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (bioMérieux, France). After drying, the target plate was loaded into the matrix-assisted laser desorption ionization-time of flight mass spectrometry-based Vitek MS system (bioMérieux, France). The mass spectral fingerprint was generated and compared automatically to the Vitek MS database version 2. Both the technician’s working time (preanalytical procedure to prepare samples) and the turnaround time (automated analytical procedure to obtain results) needed for identification of ten Aeromonas and six Salmonella isolates were recorded and compared to those from the Vitek 2 system.
Gene sequence analysis
The identification results of 73 clinical isolates of enteropathogens from the Vitek MS were arbitrated by gene sequencing.
Nucleic acid extraction
Nucleic acid was extracted from bacteria using the QIAamp MinElute Virus Spin kit (Qiagen, Germany) according to the manufacturer’s instructions.
Polymerase chain reaction
Polymerase chain reaction (PCR) and reverse transcriptase-PCR (RT-PCR) were performed to obtain specific gene sequences. Primers (see Table 1) specific for conserved regions of enteropathogens were synthesized by Sangon Biotech (China) according to the literature (5-7) and used for amplification and sequencing. Conditions for PCR and RT-PCR assays were optimized to yield gene sequences. Nucleic acid amplification with region-specific primers for Salmonella, Shigelle, C. jejuni, C. coli using 2× Tag PCR Master Mix kit (Aidlab Biotechnologies, China), and amplification with 16S rDNA universal primers for Aeromonas, Plesiomonas shigelloides, Y. enterocolitica, C. difficile, V. parahaemolyticus, V. fluvialis, and V. cholerae by the PrimeScriptTM one step RT-PCR version 2.0 Kit (Takara, Japan) were performed on a C1000 thermocycler (Bio-Rad, USA) according to the manufacturers’ instructions.

Full table
Sequencing
Sequencing was completed by a commercial service (BGI tech, China) and data were subsequently analyzed using the online BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Vitek 2 analysis
Ten Aeromonas and six Salmonella isolates were identified by the Vitek 2 system (bioMérieux, France) according to the manufacturer’s instructions. Both the technician’s working time (preanalytical procedure to prepare samples) and the turnaround time (automated analytical procedure to obtain results) needed for bacterial identification were recorded.
Results
All strains gave gene sequencing results with ≥99% similarity to published species. The identification results of strain types and 73 clinical isolates of enteropathogens from both the Vitek MS system and the sequencing method are listed in Tables 2 and 3, respectively. The genus identification results of Aeromonas isolates and Salmonella isolates by the Vitek MS system were consistent with those from gene sequence analysis. C. jejuni, C. coli, P. shigelloides, Y. enterocolitica, C. difficile, V. parahaemolyticus, V. fluvialis, and V. cholera were all correctly identified to the species level by the Vitek MS system. All Shigella isolates were identified as E. coli by the Vitek MS system, which was accompanied by a comment explaining the inability to discriminate between these two genera, and the identification result of the type strain of A. hydrophila was reported as A. hydrophila/caviae.
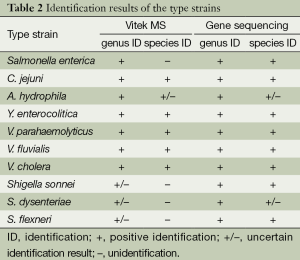
Full table
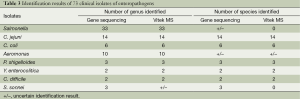
Full table
Identification of Aeromonas and Salmonella isolates by the Vitek MS system were further compared to those of the Vitek 2 system. Table 4 shows the species identification of ten Aeromonas isolates by the Vitek MS and Vitek 2 system. The identification results obtained from the Vitek MS system were essentially consistent with those from the Vitek 2 system. Furthermore, identification results of six Salmonella isolates from the Vitek MS system were exactly the same as those from the Vitek 2 system (data not listed).
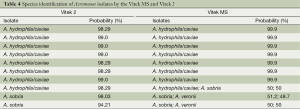
Full table
The cost of consumables and time required for identification of ten Aeromonas and six Salmonella isolates by the Vitek MS and Vitek 2 system are listed in Table 5. Consumables of the Vitek MS system include matrix solution, target plates, inoculation loops and tips. For the Vitek 2 system, the consumables include gram-negative identification cards, bacterial suspension pipes, cotton swabs and salt solution. The consumables for the Vitek MS system were in total far cheaper than those for the Vitek 2 system, and Vitek MS generated results ten times faster than the Vitek 2 system in identification of 16 isolates.

Full table
Discussion
The development of MALDI-TOF MS technology has potentially revolutionized the routine identification of microorganisms in clinical microbiology laboratories by introducing a simple, rapid, high throughput, and low-cost identification technique (3). In this study, MALDI-TOF MS is showed to be satisfactional in analytical and practical performance in identification of enteropathogens. With the exception of not being able to distinguish identification between Shigella and E.coli, MALDI-TOF MS produced correct identifications for all type strains and clinical isolates of enteropathogens representing 8 genera to the genus level. Most of the strains, including C. jejuni, C. coli, P. shigelloides, Y. enterocolitica, C. difficile, V. parahaemolyticus, V. fluvialis, V. cholera were correctly identified to the species level. Some fastidious enteropathogens such as Campylobacter used to be diagnosed by isolation of the organism on selective medium and identified manually by biochemical tests. MALDI-TOF MS especially makes identification of Campylobacter and C. difficile easier and quicker, and no longer needs large amounts of biological material and special culture environments. In addition, MALDI-TOF MS offers a clear advantage over conventional biochemical tools such as the Vitek 2, as it achieves similar analytical performance, but at lower cost and much shorter time for identification.
However, a few issues appeared in using MALDI-TOF MS for identification of enteropathogens. First, MALDI-TOF MS is unable to discriminate between E. coli and Shigella. The system reports a Shigella isolate as E. coli. The misidentification is due to the limited resolution of the MALDI-TOF MS in distinguishing between E. coli and Shigella, which are very closely related to each other and have almost identical mass spectrum. The discrimination of E. coli and Shigella will require additional tests depending on the characteristics of the isolates. Genomic studies have in fact indicated that Shigella spp. belong to the species E. coli, rather than forming a separate genus (8). Our 16S rDNA sequencing of Shigella demonstrated such similarity to E. coli (data not show). However, as Shigella causes a distinct set of disease syndromes, Shigella is still classified and reported separately from E. coli. In clinical microbiology laboratories, Shigella is preliminary distinguished from E. coli on selective agar based on special colony morphology, and then the suspected colony is further confirmed by phenotypic methods and serological tests. In general, Shigella is correctly identified in our routine practice.
Second, using MALDI-TOF MS for Salmonella typing, which is of great importance for clinical and epidemiological purposes, poses a challenge for clinical laboratories. It was reported that a larger number of reproducible peaks were required for subspecies identification, therefore, rigorous control of the sample preparation and optimization of testing parameters was critical for strain typing with MALDI-TOF MS but not practical in clinical laboratories (9). Currently, serological tests are still needed for Salmonella spp. typing. Third, although a study from Donohue MJ indicated that the mass spectral data contained sufficient information to discriminate between genera, species, and strains (10), MALDI-TOF MS sometimes produced uncertain species identification results within the Aeromonas genus. Updates to the databases may solve the problem with this species. In addition, MALDI-TOF MS cannot presently discriminate between A. hydrophila and A. caviae. This problem has been temporarily settled by identifying strains as A. hydrophila/caviae. The solution is satisfactory in the routine practice since definitive identification does not normally impact clinical management. Further defining species identification of Aeromonas and other species are hoped to be achieved with continued database development.
Conclusions
Gene sequence analysis is the “gold standard” for bacterial identification and classification while the Vitek 2 system is a popular commercial method commonly used in clinical microbiology laboratories for bacterial identification. In this study, we evaluate the analytical and practical performance of MALDI-TOF MS for identification of enteropathogens in our clinical microbiology laboratory. MALDI-TOF MS is showed to be a simple, rapid, accurate, and low-cost tool for identification of enteropathogens.
Acknowledgements
All phases of this study were supported by grant 2012ZX10004213 and 2013ZX10004805 of the National Projects of Major Infectious Disease Control and Prevention and grant 2011AA02A116 of the National High Technology Research and Development Program of China (863 Program) and grant 2009 GDUPS.
Disclosure: The authors declare no conflict of interest.
References
- Steensels D, Verhaegen J, Lagrou K. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of bacteria and yeasts in a clinical microbiological laboratory: a review. Acta Clin Belg 2011;66:267-73. [PubMed]
- Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev 2012;36:380-407. [PubMed]
- Martiny D, Busson L, Wybo I, et al. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2012;50:1313-25. [PubMed]
- Dubois D, Grare M, Prere MF, et al. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J Clin Microbiol 2012;50:2568-76. [PubMed]
- Denis M, Refrégier-Petton J, Laisney MJ, et al. Campylobacter contamination in French chicken production from farm to consumers. Use of a PCR assay for detection and identification of Campylobacter jejuni and Camp. coli. J Appl Microbiol 2001;91:255-67. [PubMed]
- Rahn K, De Grandis SA, Clarke RC, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 1992;6:271-9. [PubMed]
- Vidal M, Kruger E, Durán C, et al. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol 2005;43:5362-5. [PubMed]
- Lan R, Reeves PR. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect 2002;4:1125-32. [PubMed]
- Dieckmann R, Helmuth R, Erhard M, et al. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 2008;74:7767-78. [PubMed]
- Donohue MJ, Smallwood AW, Pfaller S, et al. The development of a matrix-assisted laser desorption/ionization mass spectrometry-based method for the protein fingerprinting and identification of Aeromonas species using whole cells. J Microbiol Methods 2006;65:380-9. [PubMed]


