Exploratory analysis of introducing next-generation sequencing-based method to treatment-naive lung cancer patients
Introduction
The treatment of cancer has revolutionized in the past decade with the development of therapies directed at specific genetic alterations, serving as a panel of druggable targets. Among all cancer types, non-small cell lung cancer (NSCLC), harboring a spectrum of mutations, has the richest targeted therapy options, ranging from compounds in clinical development to FDA-approved drugs, targeting multiple oncogenic driver genes, including epidermal growth factor receptor (EGFR) (1,2), anaplastic lymphoma kinase (ALK) (3), c-ros oncogene 1 (ROS1), ERBB2, BRAF, MET and RET (4). The characterization of NSCLC into subtypes based on their genetic alterations has significantly improved the therapeutic efficacy of targeted therapies and disease outcomes in a subgroup of patients (1,2). Accurate companion diagnosis is necessary for patient selection prior to commencing these treatment options (5-7).
Currently, single gene testing methodologies, including but not limited to ARMS-PCR, FISH and IHC are often utilized in treatment-naive patients to profile driver mutations, such as EGFR mutations and ALK fusions for classifying patients against optimal treatments (8-10). However, each of conventional technique is associated with its own disadvantages, including the low-throughput nature, limitations in detecting certain types of aberrations and the need for subjective judgments during data analysis. DNA next-generation sequencing (NGS) allows for large-scale parallel sequencing and has proved to be an effective and accurate tool for the parallel profiling of different forms of genetic abnormalities including mutations, fusions, and amplifications across a large number of genes (11). It also allows for the identification of novel mutations which cannot be identified using PCR-based methods. Countless studies have reported its utility in diagnosis and treatment guidance across a wide spectrum of cancer types. Currently in lung cancer, NGS-based methods are often performed on samples obtained from patients who have progressed on prior treatments due to the likelihood of harboring multiple mutations.
In this study, we performed capture-based ultra-deep sequencing on 61 surgically-resected samples obtained from treatment-naive patients using a panel consisting of 8 well-established driver genes for lung cancer to investigated driver mutations associated with each patient. This study evaluated the potential of utilizing NGS-based methods in treatment-naive patients and paved its way in routine clinical use in treatment-naive patients. Based on driver mutations identified using NGS, we interrogated the correlations between driver mutations and clinical characteristics in this Chinese treatment-naive cohort.
Methods
Patient selection
Between January 2016 and September 2016, 61 treatment-naive patients with resectable NSCLC were enrolled in this study. This study was approved by the Institutional Review Board at Zhangjiagang First People’s Hospital. All patients gave informed consent to participate in the study and gave permission for the use of tumor tissues.
NGS library preparation and sequencing
DNA was extracted using QIAamp DNA FFPE tissue kit (Qiagen) according to manufacturer’s instructions. DNA concentration was measured using Qubit dsDNA assay. DNA shearing was performed using Covaris M220, followed by end repair, phosphorylation and adaptor ligation. Fragments of size 200–400 bp were selected by bead (Agencourt AMPure XP Kit, Beckman Coulter, California, USA) followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. A bioanalyzer high-sensitivity DNA assay was then performed to assess the quality and size of the fragments and indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., California, USA) with pair-end reads.
Genetic profiles of all tissue samples were assessed by performing capture-based targeted deep sequencing using the 8-gene panel, covering 76 kb of human genomic regions. The 8-gene panel can detect oncogenic driver mutations of EGFR, ALK, BRAF, MET, RET, ROS1, ERBB2 and KRAS.
Sequence data analysis
Sequence data were trimmed with Trimmomatic (12) for adaptor and mapped to the human genome (hg19) using BWA aligner 0.7.10 (13). Local alignment optimization, variant calling and annotation were performed using Genome Analysis ToolKit (GATK) 3.2 (14), Picards (http://picard.sourceforge.net/) and VarScan (15). Variants were filtered using the VarScan fpfilter pipeline. At least 5 supporting reads were needed for INDELs, while 8 supporting reads were needed for SNVs to be called. According to the ExAC, 1000 Genomes, dbSNP, ESP6500SI-V2 database, variants with population frequency over 0.1% were grouped as SNP and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translocation analysis was performed using both Tophat2 and Factera (16) and CNVs were analyzed with inhouse algorithm based on sequencing depth.
Statistical analysis
The statistical analyses were performed using Software R. Statistical differences were calculated and presented using paired, two-tailed Student’s t-test in P value. For all statistical tests, P<0.05 was considered statistically significant.
Results
Patient characteristics
From January 2016 to September 2016, 61 treatment-naive patients with resectable NSCLC were enrolled in this study. The median age was 61.5 years, ranging from 25–84 years old. Thirty patients were female and 26 were male. Thirteen patients had a history of smoking and 43 patients were nonsmokers. Forty-eight cases were diagnosed as adenocarcinoma; 8 were squamous cell carcinoma (Table 1). Thirty-nine patients were classified as stage IA, accounting for 63.9% of the cohort; 5 patients were diagnosed (8.2%) at stage IB, 1 at stage II, 1 at stage IIIa, 2 at stage IIIb, and 3 patients with stage IV.
Full table
Mutation spectrum
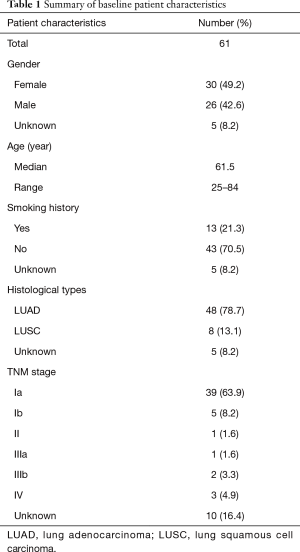
To elucidate driver mutations associated with each patient, we performed capture-based ultra-deep targeted sequencing using a panel consisting of all exons of 8 well-established driver genes for lung cancer, spanning 76 kb of human genome. We identified mutations from 80% (49/61) patients and the remaining 12 (20%) patients had no mutation identified from this panel. Collectively, 79 genetic variants were identified including 43 single nucleotide variations (SNVs), 28 insertions or deletions (INDELs), 4 copy-number amplifications (CNAs), and 4 translocations.
EGFR is the most frequently mutated gene, accounting for 44% of all variants identified (Figure 1A). Four types of EGFR mutations were detected in this study, including in-frame deletions in exon 19, single missense substitution mutation L858R, G719A, and exon 20 insertions. Of the EGFR somatic mutations, 83% of them were TKI sensitive mutations, including exon 19 deletions, L858R and G719A; 14% were EGFR-TKI resistant mutations, exon 20 insertion. Moreover, one patient was detected with EGFR G719A mutation (Figure 1B).
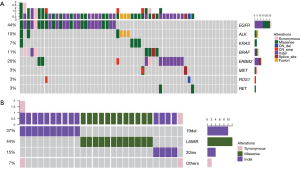
Twelve patients (20%) were identified with ERBB2 mutations. Among them, 13% of patients (8/61) harbored ERBB2 exon 20 insertion, higher than reported by previous studies, ranging from 2% to 4% (17-19). This variant located in ERBB2 tyrosine kinase domain and can affect protein function in a large extent (Figure 2).

One patient was detected with MET skipping splicing and another with MET copy number amplification. MET exon 14 skipping promotes MET RNA-splicing-based skipping, thus leading to MET kinase activity stimulation. Exon 14 encoded protein can recruit ubiquitin ligase CBL for a result of MET degradation (20-22). MET exon 14 deficiency will cause MET protein stability and persistent activation of downstream signaling (23). Taken together, exon 14 splicing alteration gives rise to oncogenic MET activation for tumorigenesis.
Seven patients carried BRAF mutations; among them, one patient harbored a rare BRAF mutation K601E, which is not included in commercially available BRAF mutations detection kit. BRAF K601E, located between two major BRAF phosphorylation sites T599 and S602 in the activation loop in the vicinity of the classic mutation V600E, has been reported in melanoma (24) and follicular thyroid carcinoma (25).
ALK fusion was found in three samples (5%), and KRAS single missense mutation G12D in four patients (6.6%). ROS1 copy number deletion and RET mutation were also detected in our cohort. In addition, 12 patients (12/61) had no mutation identified from this panel.
Of all the patients harbored driver mutation, only one patient had dual driver mutations, harboring both EGFR L858R and KRAS G12D mutation (Figure 1A), in an agreement with previous findings that driver mutations exhibit mutually exclusive correlation in treatment-naive patients (26).
Correlation between driver mutations and clinical characteristics
Next, we investigated the correlation between driver mutations and clinical parameters (including but not limited to age, gender, smoking history, histology) and expression of immunohistochemistry (IHC) markers (including but not limited to CK7, TTF-1, p40, CD56) in this Chinese treatment-naive lung cancer cohort.
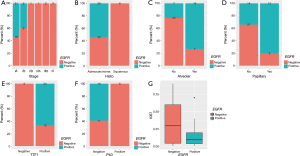
First, we interrogated the correlation of EGFR mutations and clinical features. We revealed that high prevalence of EGFR mutations was more commonly found in patients diagnosed at early-stage (P=0.0093, Figure 3A). EGFR mutation was significantly correlated with adenocarcinoma histology than squamous cell carcinoma (P=0.0052, Figure 3B). Tumors with EGFR-positive mutation were more likely to have alveolar (P=0.0004, Figure 3C) and papillary features (P=0.0053, Figure 3D) than that of EGFR-negative. As to expression of IHC markers, we observed that patients harboring EGFR mutation commonly had high expression of TTF1 (P=0.0003, Figure 3E), and low level of P40 (P=0.0184, Figure 3F) and Ki67 (P=0.0109, Figure 3G), compared to EGFR-negative patients.
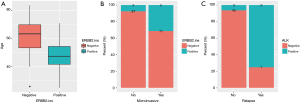
Clinical relevance of ERBB2 and ALK genomic alterations were also investigated. The alteration of ERBB2 was positively correlated with younger age (P=0.0077, Figure 4A). Tumors detected with positive-ERBB2 were significantly correlated with micro-invasive features than ERBB2-negative tumors (P=0.0351, Figure 4B). For patients harboring ALK alterations, they were more prone to relapse than patients without ALK alterations (P=0.0035, Figure 4C).
Discussion
In this study, we investigated the potential of introducing NGS-based testing methods to treatment-naive patients in a cohort of 61 treatment-naive patients by performing capture-based ultra-deep targeted sequencing using a panel consisting of 8 well-established driver genes for lung cancer. Our data revealed that 80% of patients carried driver mutation, which exhibits a mutually exclusive relationship with each other except for one case. Our data also reveal rare mutations, which can not be identified using PCR-based methods. This study highlights the potential of NGS in treatment-naive NSCLC patients. Moreover, most of previous studies were focusing on distinct molecular feature of lung cancers. Here, we clarified the spectrum of well-established oncogenic driver mutations in a Chinese treatment-naive lung cancer cohort, and analyzed the correlation between driver alterations and clinical characteristics.
Currently, single gene testing is often performed on treatment-naive patients to detect mutations to guide subsequent treatment. NGS-based methods are often used in patients who have developed resistance to prior therapies. Single gene testing methods, such as amplification refractory mutation system polymerase chain reaction (ARMS-PCR), fluorescent in situ hybridization (FISH) and IHC, are associated with a few pitfalls, including but not limited to detection of known variants, limited ability in multiplexing, and involving subjective judgement (27). Our study demonstrated that NGS-based methods allow for large-scale parallel sequencing to detect a wide range of genetic mutations simultaneously.
Using NGS, we identified numerous mutations that are not covered by commercially available testing kits, such as EGFR exon 20 insertions, ERBB2 exon 20 insertions, MET exon 14 skipping and BRAF K601E. Although the traditional method ARMS-PCR is often utilized to detect mutations in treatment-naive patients, all above mentioned mutations are not covered by commercially available kits. Thus, patients harboring such mutations may miss therapeutic opportunities if subject to test using commercially available single gene testing kits. It has been reported that patients with MET exon 14 skipping are responsive to crizotinib and positive detection of exon 14 could guide following targeted therapy for clinical benefit achievement (28). BRAF K601E located between two phosphorylation sites in BRAF activation domain (29,30). Preclinical studies reported that MEK inhibitor, trametinib (Mekinist, GSK1120212) can block the downstream signaling induced by BRAF K601E mutation (31,32), which implies that BRAF K601E mutation maybe sensitive to trametinib. Therefore, NGS should be recommended when multiple genes need to be tested.
In addition, our data revealed that 20% of patients in this cohort did not carry mutation from this panel. Several potential reasons might be responsible for this. This may attribute to tumor heterogeneity that the sampled tissue did not carry driver mutations. Another possible explanation can be the relatively low tumor content in the biopsy tissue. Furthermore, there are a significant percentage of patients who do not harbor driver mutation. Their cancer is not driven by driver mutations but another mechanisms or mutations which are not covered by our panel. Collectively, in this study, we revealed the essentialness of introducing NGS-based testing methods to treatment-naive patients.
Although NGS has been widely regarded as a powerful tool to accurately detect gene alterations in a high-throughput scale, it still has its own limitations (33,34). First, the period for NGS procedure is commonly longer for clinical diagnosis when compared to conventional methods such as ARMS-PCR and FISH. Second, the cost is an obstacle for patients to choose NGS for mutation identification. Third, the accuracy of new platform may less accurate than conventional method on specific mutation type identification. However, although challenges were created by these limitations, we should keep in mind that NGS will continue to be improved and optimized with respect to these disadvantages.
Nowadays, translational medicine has progressed to the point where patients can be stratified basing on the correlation of clinically relevance and molecular features (35,36). This kind of association might be helpful for diagnosis and patient selection for specific tumor mutations in clinical trials. There have been many studies examining the relationship between patient characteristics and oncogenic driver mutations in NSCLCs. However, several data were conflicting among different studies (37-40). Here, on the basis of oncogenic driver mutation identified by NGS, we conducted this kind of investigation in our study and tried to interrogate the underlying clinical features of tumors with different driver mutations in this treatment-naive Chinese NSCLC cohort. The presence of EGFR mutations in a subset of NSCLC were associated with a favorable prognosis in cancer patients treated with EGFR tyrosine kinase inhibitors (TKIs) (1,41). Identification of EGFR mutated-patients may indicate potential sensitive patients who would be benefit from EGFR-TKI. In this study, we found that EGFR mutations were correlated with early stage, indicating that this well-known oncogene driver mutation was acquired in early step of pulmonary carcinogenesis. Next, we evaluated whether morphological features reflect EGFR mutation status. We found that EGFR mutations were more frequently observed in tumor with alveolar or papillary components than that without such components. Our results revealed that tumors with overexpressed TTF-1 and low expressed P40 and Ki67 would have more EGFR mutations, which indicated that these immunohistological markers such as TTF-1, P40 and Ki67 can predict EGFR mutation. These results suggested that EGFR mutation test should be performed in tumors with such clinical features. Gene alterations in the ERBB2 or ALK also identify distinct subsets of NSCLC and represent a therapeutic target with already proven sensitivity to ERBB2 or ALK inhibitors in clinical settings. Here, we found ERBB2 mutations were correlated with younger age and micro-invasive potential. ALK mutations indicated further relapse probability of tumors. Although discrepancies could be introduced by the limited number of cases, these findings suggested that these clinical characteristics could be served as indicators for presence of ERBB2 and ALK mutations.
In conclusion, treatment-naive patients often undergo a few single gene tests to identify driver mutations. Several well-established driver mutations cannot be detected using commercially available single gene testing kit. We demonstrated a preferable way to profile a wide range of genetic alterations in tissue biopsy of treatment-naive NSCLC patients by NGS, highlight the needs for treatment-naive patients to undergo genomic profiling. In addition, the investigation of correlation between oncogenic driver mutations and clinical characteristics would implicate mutations examination in specific patients, which further guides more precise targeted therapy and helps patients to achieve better clinical benefit.
Acknowledgements
We thank all the patients and their families for their participation. We thank all the physicians who gave constructive comments. We thank Dr. Han Han-Zhang and Dr. Tengfei Zhang from Burning Rock Biotech for the valuable discussion and writing assistance. We also thank Dr. Ben Van der Zeijst and Dr. Peter Ujhazy for the suggestions of manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Institutional Review Board at Zhangjiagang First People’s Hospital (No. 2017003). Written informed consent was obtained from patients for this study and publication of this manuscript.
References
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015;16:e447-59. [Crossref] [PubMed]
- Mazieres J, Zalcman G, Crino L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 2015;33:992-9. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Kobayashi T, Fujimoto H, Gabazza EC. Efficacy of crizotinib in ALK fusion variants. J Thorac Dis 2016;8:E1381-3. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Farago AF, Azzoli CG. Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer. Transl Lung Cancer Res 2017;6:550-9. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Wu YC, Chang IC, Wang CL, et al. Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. PLoS One 2013;8. [Crossref] [PubMed]
- V Laffert M, Warth A, Penzel R, et al. Anaplastic lymphoma kinase (ALK) gene rearrangement in non-small cell lung cancer (NSCLC): results of a multi-centre ALK-testing. Lung Cancer 2013;81:200-6. [Crossref] [PubMed]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005;65:1642-6. [Crossref] [PubMed]
- Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 2004;431:525-6. [Crossref] [PubMed]
- Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910-8. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Lee CC, Yamada KM. Identification of a novel type of alternative splicing of a tyrosine kinase receptor. Juxtamembrane deletion of the c-met protein kinase C serine phosphorylation regulatory site. J Biol Chem 1994;269:19457-61. [PubMed]
- Lee JM, Kim B, Lee SB, et al. Cbl-independent degradation of Met: ways to avoid agonism of bivalent Met-targeting antibody. Oncogene 2014;33:34-43. [Crossref] [PubMed]
- Peschard P, Fournier TM, Lamorte L, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell 2001;8:995-1004. [Crossref] [PubMed]
- Bowyer SE, Rao AD, Lyle M, et al. Activity of trametinib in K601E and L597Q BRAF mutation-positive metastatic melanoma. Melanoma Res 2014;24:504-8. [Crossref] [PubMed]
- Pennelli G, Vianello F, Barollo S, et al. BRAF(K601E) mutation in a patient with a follicular thyroid carcinoma. Thyroid 2011;21:1393-6. [Crossref] [PubMed]
- Hsu KH, Ho CC, Hsia TC, et al. Identification of five driver gene mutations in patients with treatment-naive lung adenocarcinoma in Taiwan. PLoS One 2015;10. [Crossref] [PubMed]
- Khodakov D, Wang C, Zhang DY. Diagnostics based on nucleic acid sequence variant profiling: PCR, hybridization, and NGS approaches. Adv Drug Deliv Rev 2016;105:3-19. [Crossref] [PubMed]
- Wang SX, Zhang BM, Wakelee H, et al. PS01.67: Case Series of MET Exon 14 Skipping Mutation-Positive Non-Small Cell Lung Cancers and Response to Crizotinib: Topic: Medical Oncology. J Thorac Oncol 2016;11:S312-3. [Crossref] [PubMed]
- Vakiani E, Solit DB. KRAS and BRAF: drug targets and predictive biomarkers. J Pathol 2011;223:219-29. [Crossref] [PubMed]
- Barollo S, Pezzani R, Cristiani A, et al. Prevalence, tumorigenic role, and biochemical implications of rare BRAF alterations. Thyroid 2014;24:809-19. [Crossref] [PubMed]
- Kim KB, Kefford R, Pavlick AC, et al. Phase II study of the MEK1/MEK2 inhibitor Trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. J Clin Oncol 2013;31:482-9. [Crossref] [PubMed]
- Dahlman KB, Xia J, Hutchinson K, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2012;2:791-7. [Crossref] [PubMed]
- Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17:333-51. [Crossref] [PubMed]
- Xuan J, Yu Y, Qing T, et al. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett 2013;340:284-95. [Crossref] [PubMed]
- Fratta P, Nirmalananthan N, Masset L, et al. Correlation of clinical and molecular features in spinal bulbar muscular atrophy. Neurology 2014;82:2077-84. [Crossref] [PubMed]
- Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007;50:113-30. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]


