
MicroRNA351 targeting TRAF6 alleviates dexamethasone-induced myotube atrophy
Introduction
Glucocorticoids are a class of corticosteroids secreted from the adrenal gland, and they are usually used as therapeutic agents owing to their potent anti-inflammatory, anti-shock, and immunosuppressive functions (1-3). Dexamethasone (Dex) is commonly used in patients with malignant tumors, such as lung cancer (4,5). However, high doses and long-term use of glucocorticoids cause some passive effects including hypertension, osteoporosis, depression, skeletal muscle atrophy and so on (6-9). The molecular mechanisms of skeletal muscle atrophy have been well studied and two proteolytic pathways, the ubiquitin-proteasome system, and autophagy, have been found to play an important role in the degradation of skeletal muscle proteins. The glucocorticoid Dex promotes proliferation, protein synthesis, and degradation of C2C12 myogenic cells (8,10), and induces proteasome C3 subunit expression in L6 muscle cells (11). Therefore, glucocorticoid-treated myotubes have been widely used as an in vitro model of muscle atrophy to explore potential mechanisms.
Despite the frequent use of Dex-treated myotubes as muscle wasting experimental models to investigate possible mechanisms, numerous interesting aspects have not well elucidated. Tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) is a member of the TRAF family of conserved adaptor proteins that have been shown to be involved in the activation of various signaling pathways, including nuclear factor (NF)-kB, mitogen-activated protein kinase (MAPK), and phosphatidylinositide 3-kinase/Akt (12-17). TRAF6 is different from other TRAF family members because it has been shown to have E3 ubiquitin ligase activity and is upregulated in skeletal muscle in response to denervation, starvation, and cancer cachexia (15,18,19). Skeletal muscle-specific deletion of TRAF6 in mice results in partial sparing of muscle mass following denervation, starvation and cancer cachexia. The data suggest that TRAF6 inhibition could rescue some catabolism-induced muscle atrophy.
Recently, microRNAs (miRNAs) have been shown to participate in regulating a variety of signal pathways in the skeletal muscle, which suggests a potential association with muscle catabolism. MiRNAs act by targeting sequences in the 3ʹ untranslated region of mRNAs to increase their degradation or inhibit their translation, which eventually limits the expression of critical proteins (20). MiRNAs specifically expressed in muscles can change diseases affecting the muscle. For example, muscle-specific microRNA1 is induced during Dex-mediated muscle atrophy, whereas heat shock protein 70 (HSP70) levels are reduced (21). It appears that the expression of miR-206 is increased during the differentiation of satellite cells while miR-29 improves muscle cell proliferation (22,23). Wada et al. (24,25) demonstrated that miR-23a repressed the translation of muscle-RING-finger protein-1 (MuRF1) and atrogin-1 mRNAs; moreover, miR-23a was suppressed during diabetes and Dex-induced muscle atrophy (25).
In this study, we obtained evidence showing that the expression of miRNA-351 was downregulated and that of TRAF6 was upregulated during Dex-induced C2C12 myotube atrophy. MicroRNA351 (miR-351) can directly target the 3'UTR of Traf6. Overexpression of miRNA-351 by mimic transfection alleviated Dex-mediated myotube atrophy and inhibited the expression of TRAF6, MuRF1, and muscle atrophy F-box (MAFbx).
Methods
Cell culture and transfection
Mouse C2C12 myoblasts (Cell Bank, Chinese Academy of Sciences, Shanghai, China) were cultured in growth media [Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-BRL, Gaithersburg, MD, USA) plus 10% fetal bovine serum (FBS) (Gibco-BRL, Gaithersburg, MD, USA), 100 U/mL penicillin (Sigma-Aldrich, St. Louis, MO, USA), and 100 µg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA)]. Myoblasts were grown to approximately 90% confluence in a six-well plate and then differentiated into myotubes by replacing the growth media with differentiation media (DMEM supplemented with 2% FBS and 1% penicillin and streptomycin) for 6 days. Then, the formed C2C12 myotubes were treated with 100 nM Dex in 0.1% ethanol for 24 and 48 h while C2C12 myotubes cultured in the vehicle (0.1% ethanol-containing medium) were used as the control.
The myotubes were transfected with 40 nM miR-351 mimic, negative control, or 100 nM miR-351 inhibitor using Lipofectamine 2000 (Invitrogen). After 6 h, the medium of the transfected myotubes was switched to atrophy medium for 24 or 48 h as described above. After treatments, the transfected C2C12 myotubes were photographed under a phase contrast microscope (Leica Microsystems, Wetzlar, Germany). The diameter of C2C12 myotubes was determined at three points along the length of each myotube in a blinded fashion, and the average diameter per myotube was expressed as the mean of three measurements. At least 50 myotubes were measured using the Image-Pro Plus software (Media Cybernetics, Silver Springs, MD, USA).
Real-time reverse transcription-polymerase chain reaction (RT-qPCR)
The RNA samples were reverse transcribed using the QuantiNova Reverse Transcription (RT) kit (QIAGEN, 205411) while the miR-351 expression was measured using the miScript II RT kit (QIAGEN, 218161). The following primers used in this study were prepared by the Shanghai Generay Biotech Co., Ltd., (Shanghai, China): Traf6: forward (F), GCAGAGGAATCACTTGGCACG and reverse (R), CACGGACGCAAAGCAAGGTT; hypoxanthine phosphoribosyltransferase 1 (HPRT1): F, AGTCCCAGCGTCGTGATTAGC and R, GTGATGGCCTCCCATCTCCTT; miR-351-5PI: F, ACACTCCAGCTGGGTCCCTGAGGAGCCCTTTG and R, CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGGCTC; TRP sequence: TGGTGTCGTGGAGTCG; and U6: F, CTCGCTTCGGCAGCACA and R, AACGCTTCACGAATTTGCGT. Samples were measured in triplicate and normalized to U6, which has frequently been used as control RNA for miRNA in C2C12 cells. To quantify TRAF6 levels, HPRT1 was used as a normalization control. All RT-qPCR experiments were performed using the QuantiNova SYBR Green PCR kit (QIAGEN, 208054) on the following cycling schedule: 95 °C for 2 min and 40 cycles of 95 °C for 15 sec and 60 °C for 1 min. The relative expression was measured using the 2−ΔΔCt method.
Western blot analysis
For the western blot analysis, cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Haimen, China). Then, the total proteins were separated using electrophoresis, transferred to a polyvinylidene fluoride (PVDF), which was blocked in 5% skim milk, and then incubated at 4 °C overnight with the following primary antibodies: rabbit anti-TRAF6 polyclonal (1:1,000, ABGENT, San Diego, CA, USA), goat anti-MuRF1 polyclonal (1:1,000, R&D Systems, Minneapolis, MN, USA), anti-MAFbx polyclonal (1:1,000, LifeSpan Biosciences, Seattle, WA, USA), and rabbit anti-beta tubulin polyclonal (1:2000, Abcam, Cambridge, MA, USA). The next day, the PVDF membranes were washed with Tris-buffered saline with Tween (TBST), incubated with the corresponding secondary antibody at room temperature for 1 h, washed with TBST three times, and exposed to visualization reagents followed by quantification using the ImageJ software program.
Statistical analysis
All the data were expressed as means ± standard error of the mean (SEM) as specifically indicated. A one-way analysis of variance (ANOVA) followed by Dunnett’s test was used to compare differences among groups.
Results
TRAF6 and miR-351 were inversely related in Dex-treated C2C12 myotubes
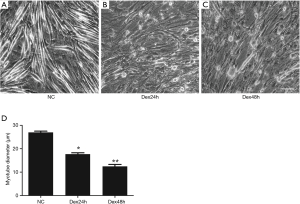
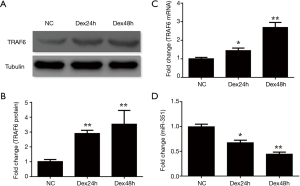
We investigated the expression of miRNA-351 and TRAF6 during Dex-induced myotube atrophy using C2C12 cells. Results showed that the myotube morphology became irregular and the diameter of the C2C12 myotubes decreased 24 and 48 h after Dex treatment (Figure 1A,B,C,D). The expression of TRAF6 mRNA and protein was significantly increased compared to that of the negative control group at different time points (Figure 2A,B,C), whereas the expression of miR-351 was reduced (Figure 2D). These data indicate that the expression of miR-351 and TRAF6 was inversely related during Dex-induced myotube atrophy.
miR-351 interacted with 3'UTR of TRAF6
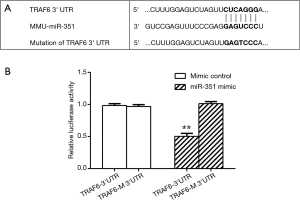
We sought to determine whether miR-351 could be directly involved in suppressing the predicted target gene, TRAF6, by cloning its 3'UTR into a luciferase reporter construct, which was transfected into 293T cells with plasmid constructs overexpressing either miR-351 or scrambled miRNA control. The 3'UTR of TRAF6 was predicted to have a single miR-351 target site essential for miRNA/mRNA binding (Figure 3A). The luciferase reporter analysis showed that the miR-351 mimic control did not affect luciferase activity in both the TRAF6 3'UTR and mutated TRAF6 3'UTR (TRAF6-M 3'UTR) groups. Interestingly, the miR-351 mimic suppressed luciferase activity in the TRAF6 3'UTR group but not the TRAF6-M 3'UTR group (Figure 3B). These data showed an interaction between miR-351 and 3'UTR of TRAF6.
miR-351 alleviated C2C12 myotube atrophy by targeting TRAF6
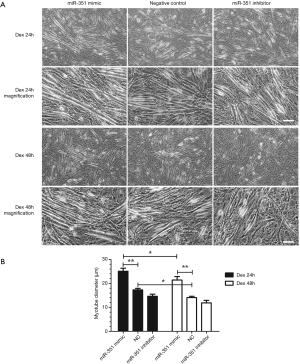
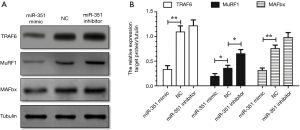
We investigated the effects of Dex on miR-351 mimics in the presence of inhibitors on transfected C2C12 myotubes. Results showed that compared with the negative control group, the diameter of C2C12 myotubes in the miR-351 mimic transfection group was significantly increased. However, the diameter of C2C12 myotubes in the miR-351 inhibitor group was significantly reduced (Figure 4). This result suggests that miR-351 relieved the Dex-induced muscle atrophy. The results of the western blot analysis showed that the expression of TRAF6 was significantly decreased and accompanied by a decline in the expression of MuRF1 and MAFbx in the miR-351 mimic transfection group (Figure 5). This finding demonstrates that miR-351 inhibited the expression of TRAF6, MuRF1, and MAFbx in Dex-treated C2C12 myotubes. At the same time, TRAF6, MuRF1, and MAFbx were all upregulated in the miR-351 inhibitor transfection group, but the difference was not significant, except for that of MuRF1.
Discussion
Glucocorticoids, a class of steroid hormones secreted by the adrenal cortex, exert immunosuppressive, anti-inflammatory, and anti-shock effects (26). They are commonly used to clinically treat many diseases. However, long-term use of glucocorticoids produces certain adverse effects such as muscle atrophy (23,27). Glucocorticoids had been known to activate muscle atrophy through the transcriptional induction of two E3 ubiquitin ligases, MuRF1 and MAFbx, as well as activation of proteasome degradation (8,19,28). Moreover, the expression of TRAF6 was upregulated and induced muscle atrophy by activating MuRF1 and MAFbx under denervation conditions (6,8,29).
In this study, C2C12 myotubes were used for Dex-induced muscle atrophy model. We found that the expression of TRAF6 was increased, which was in agreement with our previous results (8). In addition, we found that the expression of miR-351 was downregulated in the Dex-treated C2C12 myotubes. Our previous study showed that miR-351 was downregulated in denervation-induced skeletal muscle atrophy (6), which was consistent with and confirmed current results. Based on the Targetscan analysis, a number of miRNAs may target TRAF6 including miR-351. Furthermore, our results showed that the expression of miR-351 and TRAF6 was inversely related during Dex-induced C2C12 myotube atrophy. Therefore, miR-351 was chosen for detailed investigation, and the results showed that miR-351 directly targeted TRAF6 3'UTR.
Next, we found that transfection with miR-351 prevented the Dex-induced upregulation of TRAF6 that was crucial for increasing the activity of the proteolysis systems (UPS and ALS) in C2C12 myotubes (30). These data were consistent with those of previous studies indicating that miR-351 was downregulated in denervated skeletal muscle (6,8). MuRF1 and MAFBx are well-known downstream signaling molecules of TRAF6 (18) and, therefore, their expression was determined and found to be upregulated in Dex-induced C2C12 myotube atrophy. Transfection of miR-351 abrogated the reduction of the C2C12 myotube diameter induced by Dex, and the increased expression of MuRF1 and MAFBx, which are required to enhance the activity of the ubiquitin-proteasome system. These data were in line with the in vivo results (6,8) and suggested that miR-351 inhibited the expression of TRAF6, MuRF1, and MAFbx in Dex-treated C2C12 myotubes. Du et al. have found that miR-351-5p overexpression promoted the proliferation and inhibited the differentiation of C2C12 myoblast, as well as mediated the regulation of muscle fiber type transition in vivo (31). A recent study has demonstrated that miR-351 aggravates intestinal ischaemia/reperfusion injury through the targeting of MAPK13 and Sirtuin-6 (32). Intriguingly, it has been showed that TRAF6, an adaptor protein which functions as an E3 ubiquitin ligase, is an important regulator of satellite cell homeostasis in adult skeletal muscle (33). Lack of TRAF6 has some influences in the expression levels of miR-1, miR-133, miR-206 in cultured myogenic cells. It has been indeed identified that a network of myomiRs regulates the expression of genes involved in regulating muscle structures and functions during myogenesis or atrophy (34). In our study, we provided further evidence for the involvement of miR-351 targeting TRAF6 in muscle atrophy and contributed to the growing evidence that miRNAs, including miR-351, regulate muscle atrophy through posttranscriptional mechanism under a variety of catabolic conditions.
In summary, this study showed that the expression of miR-351 and TRAF6 was inversely related during Dex-induced C2C12 myotube atrophy. We also noted that treatment with miR-351 significantly inhibited the Dex-induced C2C12 myotube atrophy by suppressing the expression of TRAF6, MuRF1, and MAFbx. Collectively, our results provided further evidence for the involvement of miR-351 targeting TRAF6 in muscle atrophy and contributed to the growing evidence that miRNAs, including miR-351, regulate muscle atrophy through posttranscriptional mechanism under a variety of catabolic conditions.
Acknowledgements
Funding: This study was supported by the National Key Research and Development Program of China (Grant No. 2017YFA0104703), the National Natural Science Foundation of China (Grants No. 81671230, 81871554 and 81301628), the 973 Program (Grant No. 2014CB542202 and 2014CB542203), a project funded by Jiangsu Provincial Key Medical Center, the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), China Postdoctoral Science Foundation (Grant No. 2016M591894), Jiangsu Postdoctoral Science Foundation (Grant No. 1601040A), and Fund of Doctoral Start-up of Nantong University (Grant No. 15B18 and 17ZZ040).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Zelena D, Makara BG. Steroids: The physiologic and pharmacologic effects of glucocorticoids. Orv Hetil 2015;156:1415-25. [Crossref] [PubMed]
- Malkawi AK, Alzoubi KH, Jacob M, et al. Metabolomics Based Profiling of Dexamethasone Side Effects in Rats. Front Pharmacol 2018;9:46. [Crossref] [PubMed]
- Zhao SQ, Xu SQ, Cheng J, et al. Anti-inflammatory effect of external use of escin on cutaneous inflammation: possible involvement of glucocorticoids receptor. Chin J Nat Med 2018;16:105-12. [PubMed]
- Mulvenna P, Nankivell M, Barton R, et al. Dexamethasone and supportive care with or without whole brain radiotherapy in treating patients with non-small cell lung cancer with brain metastases unsuitable for resection or stereotactic radiotherapy (QUARTZ): results from a phase 3, non-inferiority, randomised trial. Lancet 2016;388:2004-14. [Crossref] [PubMed]
- Saito M, Kiyozaki H, Obitsu T, et al. Herpes simplex virus-1 encephalitis induced by chemoradiotherapy and steroids in an esophageal cancer patient: a case report. BMC Cancer 2016;16:233. [Crossref] [PubMed]
- He Q, Qiu J, Dai M, et al. MicroRNA-351 inhibits denervation-induced muscle atrophy by targeting TRAF6. Exp Ther Med 2016;12:4029-34. [Crossref] [PubMed]
- Jesinkey SR, Korrapati MC, Rasbach KA, et al. Atomoxetine prevents dexamethasone-induced skeletal muscle atrophy in mice. J Pharmacol Exp Ther 2014;351:663-73. [Crossref] [PubMed]
- Sun H, Gong Y, Qiu J, et al. TRAF6 inhibition rescues dexamethasone-induced muscle atrophy. Int J Mol Sci 2014;15:11126-41. [Crossref] [PubMed]
- Horowitz MA, Zunszain PA, Anacker C, et al. Glucocorticoids and inflammation: a double-headed sword in depression? How do neuroendocrine and inflammatory pathways interact during stress to contribute to the pathogenesis of depression? Mod Trends Pharmacopsychiatry 2013;28:127-43. [Crossref] [PubMed]
- Desler MM, Jones SJ, Smith CW, et al. Effects of dexamethasone and anabolic agents on proliferation and protein synthesis and degradation in C2C12 myogenic cells. J Anim Sci 1996;74:1265-73. [Crossref] [PubMed]
- Du J, Mitch WE, Wang X, et al. Glucocorticoids induce proteasome C3 subunit expression in L6 muscle cells by opposing the suppression of its transcription by NF-kappa B. J Biol Chem 2000;275:19661-6. [Crossref] [PubMed]
- Ma W, Xu T, Wang Y, et al. The role of inflammatory factors in skeletal muscle injury. Biotarget 2018;2:7. [Crossref]
- Lamothe B, Besse A, Campos AD, et al. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem 2007;282:4102-12. [Crossref] [PubMed]
- Paul PK, Gupta SK, Bhatnagar S, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol 2010;191:1395-411. [Crossref] [PubMed]
- Yang WL, Wang J, Chan CH, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 2009;325:1134-8. [Crossref] [PubMed]
- Takaesu G. Two types of TRAF6-dependent TAK1 activation in the IL-1 signaling pathway. Biotarget 2018;2:2. [Crossref]
- Huang Z, Zhu J, Ma W, et al. Strategies and potential therapeutic agents to counter skeletal muscle atrophy. Biotarget 2018;2:8. [Crossref]
- Paul PK, Bhatnagar S, Mishra V, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol 2012;32:1248-59. [Crossref] [PubMed]
- Qiu J, Fang Q, Xu T, et al. Mechanistic Role of Reactive Oxygen Species and Therapeutic Potential of Antioxidants in Denervation- or Fasting-Induced Skeletal Muscle Atrophy. Front Physiol 2018;9:215. [Crossref] [PubMed]
- Fang Q, Xu T, Wu C, et al. Biotargets in Neural Regeneration. Biotarget 2017;1:6. [Crossref]
- Kukreti H, Amuthavalli K, Harikumar A, et al. Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J Biol Chem 2013;288:6663-78. [Crossref] [PubMed]
- Nakasa T, Ishikawa M, Shi M, et al. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med 2010;14:2495-505. [Crossref] [PubMed]
- Li J, Chan MC, Yu Y, et al. miR-29b contributes to multiple types of muscle atrophy. Nat Commun 2017;8:15201. [Crossref] [PubMed]
- Wada S, Kato Y, Okutsu M, et al. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem 2011;286:38456-65. [Crossref] [PubMed]
- Hudson MB, Woodworth-Hobbs ME, Zheng B, et al. miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol 2014;306:C551-8. [Crossref] [PubMed]
- Jiang CL, Liu L, Li Z, et al. The novel strategy of glucocorticoid drug development via targeting nongenomic mechanisms. Steroids 2015;102:27-31. [Crossref] [PubMed]
- Bhatnagar S, Mittal A, Gupta SK, et al. TWEAK causes myotube atrophy through coordinated activation of ubiquitin-proteasome system, autophagy, and caspases. J Cell Physiol 2012;227:1042-51. [Crossref] [PubMed]
- Schakman O, Kalista S, Barbe C, et al. Glucocorticoid-induced skeletal muscle atrophy. Int J Biochem Cell Biol 2013;45:2163-72. [Crossref] [PubMed]
- Hindi SM, Sato S, Choi Y, et al. Distinct roles of TRAF6 at early and late stages of muscle pathology in the mdx model of Duchenne muscular dystrophy. Hum Mol Genet 2014;23:1492-505. [Crossref] [PubMed]
- Paul PK, Kumar A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy 2011;7:555-6. [Crossref] [PubMed]
- Du J, Zhang P, Zhao X, et al. MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-beta and is regulated by lnc-mg. FASEB J 2018:fj201701394RRR.
- Hu Y, Tao X, Han X, et al. MicroRNA-351-5p aggravates intestinal ischaemia/reperfusion injury through the targeting of MAPK13 and Sirtuin-6. Br J Pharmacol 2018;175:3594-609. [Crossref] [PubMed]
- Hindi SM, Kumar A. TRAF6 regulates satellite stem cell self-renewal and function during regenerative myogenesis. J Clin Invest 2016;126:151-68. [Crossref] [PubMed]
- McCarthy JJ. The MyomiR network in skeletal muscle plasticity. Exerc Sport Sci Rev 2011;39:150-4. [Crossref] [PubMed]


