
The methylation induced by protein arginine methyltransferase 5 promotes tumorigenesis and progression of lung cancer
Introduction
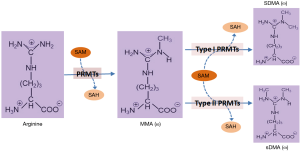
Post-translational modification (PTM) is involved in regulating many biological processes. The methylation of arginine residues is a common form of PTM, which is catalyzed by a distinct group of transmethylases known as the protein arginine methyltransferases (PRMTs). PRMTs widely exist in eukaryotic cells and participate in epigenetic regulation through the methylation of arginine residues of histone and/or transcription factors. In particular, such methylation modifications are performed by PRMTs, which catalyze and transfer methyl group (CH3) from S-adenosylmethionine (SAM) to the guanidine nitrogen of arginine residues. At present, PRMTs are classified into two subgroups according to the types of methylation they catalyze (Figure 1), namely type I and type II. The former one includes PRMT1, 2, 3, 4, 6, and 8, catalyze the formation of ω-NG-monomethyl arginine (MMA) and ω-NG, NG-asymmetric dimethylarginine (aDMA) (1-5), while the latter one covers PRMT5, 7, and 9, generate MMA and ω-NG, N'G-symmetric dimethylarginine (sDMA) residues (6). With each addition of CH3 to the guanidine nitrogen of arginine residues, a potential hydrogen bond donor would be removed, which further alters the structure of the target proteins. Both aDMA and sDMA, for example, can diversify the hydrophobicity and bulkiness of a target protein, which accordingly impact the protein-protein interaction, RNA processing, DNA repair, transduction of intracellular signaling, and regulation of gene expression.
As one of the most typical representatives of symmetric arginine methyltransferases, both the complete loss and deactivation of PRMT5 can abrogate the growth and development of cancer cells (7,8). Typically, PRMT5 is abnormally overexpressed in cancer cells and involved in a variety of crucial cellular processes that regulate proliferation, differentiation, growth, invasion and metastasis of several kinds of cancer cells. It indicates that PRMT5 is implicated in the pathogenesis and progression of various cancers. Insights into the effects of PRMT5 on tumorigenesis and progression of lung cancer are expected to provide new approaches for the prevention and diagnosis of lung cancer. In addition, it is necessary to emphasize the evidence that links PRMT5-induced methylation with lung cancer progression.
Overview of PRMT5
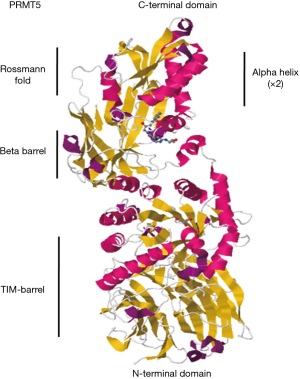
PRMT5, as a human homolog of Hsl7 (Saccharomyces cerevisiae) and Skb1 (Schizosaccharomyces pombe), was initially identified as an interacting protein of Janus tyrosine kinase 2 (Jak2), and accordingly, it is usually referred to as Jak-binding protein 1 (JBP1). PRMT5 is located in human chromosome 14q11.2, with its upstream genes of Jub, c14 and f94 and downstream genes of RNA binding motif protein 23 (RBM23), and its structure sequence is highly conservative. What is more, the C-terminal of PRMT5 contains one classic Rossmann fold and two alpha helixes, both of which are the binding sites of SAM (also known as AdoMet). These special domains are assembled with the barrel structure of N-terminal and further form a slit structure which provides options to bind with protein substrate specifically (Figure 2). Histones are the major substrates of PRMT5 and the methylation has a significant effect on the expression of the downstream genes. Furthermore, the PRMT5-driven arginine methylation of histone H2a (H2aR3me2s), H4 (H4R3me2s) and H3 (H3R8me2s, H3R2me2s) can alter chromatin structure and induce transcriptional repression. The methylation of H4R3 and H3R8 induced by PRMT5 are usually considered to be correlated with repression of transcription (9-12), whereas H3R8me2s methylated by PRMT5 is occasionally related to transcriptional activation (10). Through the transcriptional regulation, PRMT5 can participate in controlling various cellular processes, such as proliferation, differentiation, invasion, migration, apoptosis, etc.
PRMT5 expression and subcellular localization
The expression and sub-cellular localization of PRMT5 are various in different cells and at different stages. PRMT5 has been reported to be overexpressed in several cancer cells, including lung cancer (8,12), prostate cancer (13), breast cancer (14,15), lymphoma (16), and myeloid leukemia (10,17,18). PRMT5 was not only detected in the nucleus during the early development of mouse, but also observed in the cytoplasm of the inner cells by embryonic day 6.5 (7,19).
In addition to the findings above, PRMT5 was found in both the nucleus and the cytoplasm in mammalian cells. In the cytoplasm, PRMT5 binds with pICln and Sm protein to form a complex, which significantly increased the integration with the survival motor neuron (20). PRMT5 also could form a complex with p44 and pICln with distinct substrate specificity, which methylated SmD3 substrate (13). In the nucleus, PRMT5 appears to form SWI/SNF and NURD chromatin remodeling complexes. In addition, there are significant differences in the sub-cellular localization of PRMT5 between tumor and normal tissues and between cancer subtypes. PRMT5 was observed in the cytoplasm in prostate cancer cells, whereas in benign prostate epithelium cells (13), it was mainly detected in the nucleus. In lung cancer, with the increase of malignancy, the total level of PRMT5 was increased, followed by the gradual increase of the corresponding cytoplasm staining (21). Up-regulation of PRMT5 was mainly located in cytoplasm in non-small cell lung cancer (NSCLC) and was closely related to the poorer outcome, whereas raised nuclear PRMT5 most often occurs in well differentiated cancer instead of poorly differentiated small cell lung cancer and large cell neuroendocrine carcinoma (22). We previously reported that PRMT5 contains three nuclear exclusion signals (NES, including PRMT5 amino acid residues: 1–90, 500–560, 576–637), which mediated the transformation of PRMT5 subcellular localization (13). Distinct types of subcellular localization of PRMT5 during tumorigenesis play different roles. The nuclear PRMT5 suppresses growth of prostate cancer cells, while the cytoplasmic PRMT5 promotes prostate cancer cell growth via methylating substrates. According to these studies, the transformation of PRMT5 subcellular localization is indicated to occur during tumorigenesis, and different subcellular localization of PRMT5 may have cell specificity, or show that it is at different stages of cellular biological function.
PRMT5-induced methylation modulates lung cancer progression
Overexpression of PRMT5 is a prognostic biomarker in lung cancer and is positively related to clinical stages, lymphatic metastasis and survival (12). Mechanistic observation on this upregulated expression in lung cancer was carried out and reported by several relative studies, in which overexpression of PRMT5 was directly related to the poor prognosis, possibly mediated by means of the miR-99 family/FGFR3 signaling axis (12), symmetric methylation of H4R3 (22) and/or epithelial-to-mesenchymal transition (EMT) (21,23). PRMT5 not only promotes lung cancer cells growth, but also facilitates lung cancer metastasis. Conversely, its inhibition and loss could disrupt cancer cell phenotypes, and lung cancer cell proliferation can be directly inhibited by silencing or inhibiting PRMT5 expression, whereas overexpression of PRMT5 is associated with cellular hyperproliferation (8,12). Moreover, both overexpression and methylation activity enhancement of PRMT5 have been proved to be closely related to lung cancer progression.
PRMT5 modulates lung cancer progression primarily through the methylation of histone or non-histone substrates. The methylation of epidermal growth factor receptors (EGFR) catalyzed by PRMT5 decreased its auto-phosphorylation, weakening its activity and regulated cell proliferation and growth (24). PRMT5 also directly repressed the transcription of the miR-99 family (miR-99a, miR-99b, and miR-100) by using symmetrical dimethylation of histone H4R3 (H4R3me2s). This transcription inhibition increased the FGFR3 expression and in turn activated Erk1/2 and Akt, leading to cell growth and metastasis of lung cancer (8,12). Additionally, knockdown of PRMT5 resulted in a significant loss of the growth and metastatic potentials of lung cancer cells. This phenotypic regulation was mediated through the EMT response of TGF-β treatment, which dramatically increased the relative enrichment of PRMT5 protein and changed EMT related protein markers with decrease of E-cadherin and increase of Vimentin and Snail. Meanwhile, the treatment of TGF-β caused an accumulation of H4R3me2s and H3R2me2s (23). Furthermore, with the help of Ajuba (a bridging molecule), PRMT5 interacted with Snail, which in turn repressed the expression of E-cadherin, resulting in the EMT process (25). Besides, PRMT5 modulated cancer progression through the regulation of gene p53, p65, E2F-1, enolase-1 (Eno-1), eukaryotic translation initiation factor (eIF4E), programmed cell death 4 (PDCD4), transcription of involucrin (hINV) Krüppel-like factor 4 (KLF4, GKLF) and microphthalmia-associated transcription factor (MITF) (14,15,26-31).
In addition, the modulation of cell cycle progression induced by PRMT5 was involved in tumor growth and metastasis (8,12,31,32). Silencing the expression of PRMT5 attenuated the cell cycle in NIH3T3 cells and arrested cell cycle at G1 phase in 293T, MCF7 and A549 cells, and eventually induced necrosis (8,27,33), It also resulted in an increase of p27 (Kip1), which inhibited the cell cycle progression (30). On the contrary, upregulation of PRMT5 increased the expression of the positive regulators of G1 phase, such as cyclin D1, cyclin D2, cyclin E1, CDK4, and CDK6, and simultaneously reduced the expression of related negative regulator of G1 phase, such as Rb protein (33). Consequently, as manifested by these results, the slowed growth of cancer cells induced by PRMT5 knockdown was partially attributed to the arrest of cell cycle at the G1 phase.
Conclusions
To sum up, we laid the emphasis on the arginine methylation of histone introduced by PRMT5 and highlighted its growth and metastasis promoting effects. PRMT5 had been demonstrated to be vital for the growth and metastasis of lung cancer, and was closely correlated with clinical stages, lymph nodes metastasis and poor survival in lung cancer patients. Overexpression of PRMT5 promoted metastasis and indicated poor prognosis of lung cancer, while both down-regulation of PTMT5 and inhibition of its methyltransferase activity resulted in slow growth and decreased metastasis. Consequently, PRMT5, as a potential biomarker for identifying lung cancer with more aggressiveness, will be very meaningful. It also raised an interesting possibility that targeting PRMT5 may inhibit both growth and metastasis potentials of lung cancer. Currently, several small molecule inhibitors targeting PRMT5, such as EPZ015666 (GSK3235025) (34,35), C9 (36), HLCL61 (10) and PJ-68 (18) had been developed or validated (35,37-39). Although there were no clinical applications for these inhibitors, several studies had expounded that they play important roles in the growth inhibition of lung cancer. The further evaluation of these inhibitors for lung cancer treatment is necessary and expected to be carried out imminently.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China (No. 81372510 and 81171922).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lin WJ, Gary JD, Yang MC, et al. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem 1996;271:15034-44. [Crossref] [PubMed]
- Yildirim AO, Bulau P, Zakrzewicz D, et al. Increased protein arginine methylation in chronic hypoxia: role of protein arginine methyltransferases. Am J Respir Cell Mol Biol 2006;35:436-43. [Crossref] [PubMed]
- Tang J, Gary JD, Clarke S, et al. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem 1998;273:16935-45. [Crossref] [PubMed]
- Chen D, Ma H, Hong H, et al. Regulation of transcription by a protein methyltransferase. Science 1999;284:2174-7. [Crossref] [PubMed]
- Frankel A, Yadav N, Lee J, et al. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem 2002;277:3537-43. [Crossref] [PubMed]
- Morales Y, Caceres T, May K, et al. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch Biochem Biophys 2016;590:138-52. [Crossref] [PubMed]
- Tee WW, Pardo M, Theunissen TW, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev 2010;24:2772-7. [Crossref] [PubMed]
- Gu Z, Gao S, Zhang F, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J 2012;446:235-41. [Crossref] [PubMed]
- Blanc RS, Richard S. Arginine Methylation: The Coming of Age. Mol Cell 2017;65:8-24. [Crossref] [PubMed]
- Tarighat SS, Santhanam R, Frankhouser D, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 2016;30:789-99. [Crossref] [PubMed]
- Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci 2015;72:2041-59. [Crossref] [PubMed]
- Jing P, Zhao N, Ye M, et al. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett 2018;427:38-48. [Crossref] [PubMed]
- Gu Z, Li Y, Lee P, et al. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One 2012;7:e44033. [Crossref] [PubMed]
- Hu D, Gur M, Zhou Z, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun 2015;6:8419. [Crossref] [PubMed]
- Powers MA, Fay MM, Factor RE, et al. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res 2011;71:5579-87. [Crossref] [PubMed]
- Koh CM, Bezzi M, Low DH, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015;523:96-100. [Crossref] [PubMed]
- Kaushik S, Liu F, Veazey KJ, et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 2018;32:499-509. [Crossref] [PubMed]
- Jin Y, Zhou J, Xu F, et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest 2016;126:3961-80. [Crossref] [PubMed]
- Bezzi M, Teo SX, Muller J, et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev 2013;27:1903-16. [Crossref] [PubMed]
- Friesen WJ, Massenet S, Paushkin S, et al. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 2001;7:1111-7. [Crossref] [PubMed]
- Ibrahim R, Matsubara D, Osman W, et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum Pathol 2014;45:1397-405. [Crossref] [PubMed]
- Shilo K, Wu X, Sharma S, et al. Cellular localization of protein arginine methyltransferase-5 correlates with grade of lung tumors. Diagn Pathol 2013;8:201. [Crossref] [PubMed]
- Chen H, Lorton B, Gupta V, et al. A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 2017;36:373-86. [Crossref] [PubMed]
- Hsu JM, Chen CT, Chou CK, et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol 2011;13:174-81. [Crossref] [PubMed]
- Hou Z, Peng H, Ayyanathan K, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 2008;28:3198-207. [Crossref] [PubMed]
- Cho EC, Zheng S, Munro S, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J 2012;31:1785-97. [Crossref] [PubMed]
- Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res 2009;37:4965-76. [Crossref] [PubMed]
- Saha K, Adhikary G, Eckert RL. MEP50/PRMT5 Reduces Gene Expression by Histone Arginine Methylation and this Is Reversed by PKCdelta/p38delta Signaling. J Invest Dermatol 2016;136:214-24. [Crossref] [PubMed]
- Wei H, Wang B, Miyagi M, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc Natl Acad Sci U S A 2013;110:13516-21. [Crossref] [PubMed]
- Nicholas C, Yang J, Peters SB, et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.). PLoS One 2013;8:e74710. [Crossref] [PubMed]
- Zakrzewicz D, Didiasova M, Kruger M, et al. Protein arginine methyltransferase 5 mediates enolase-1 cell surface trafficking in human lung adenocarcinoma cells. Biochim Biophys Acta Mol Basis Dis 2018;1864:1816-27. [Crossref] [PubMed]
- Sheng X, Wang Z. Protein arginine methyltransferase 5 regulates multiple signaling pathways to promote lung cancer cell proliferation. BMC Cancer 2016;16:567. [Crossref] [PubMed]
- Wei TY, Juan CC, Hisa JY, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci 2012;103:1640-50. [Crossref] [PubMed]
- Duncan KW, Rioux N, Boriack-Sjodin PA, et al. Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666. ACS Med Chem Lett 2015;7:162-6. [Crossref] [PubMed]
- Fuhrmann J, Clancy KW, Thompson PR. Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev 2015;115:5413-61. [Crossref] [PubMed]
- Kong GM, Yu M, Gu Z, et al. Selective small-chemical inhibitors of protein arginine methyltransferase 5 with anti-lung cancer activity. PLoS One 2017;12:e0181601. [Crossref] [PubMed]
- Kaniskan HÜ , Konze KD, Jin J. Selective Inhibitors of Protein Methyltransferases. J Med Chem 2015;58:1596-629. [Crossref] [PubMed]
- Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 2009;8:724-32. [Crossref] [PubMed]
- Dong Y , Song C, Wang YT, et al. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell Signal 2017;34:55-65. [Crossref] [PubMed]


