
Circulating free tumor DNA in non-small cell lung cancer (NSCLC): clinical application and future perspectives
Introduction
Over the past decade, molecular characterization of non-small cell lung cancer (NSCLC) has uncovered molecularly defined subsets of tumors (1,2). Somatic molecular alterations in NSCLC can lead to oncogene activation through multiple mechanisms, including point mutations, insertions, deletions and gene rearrangements. For a subset of patient, the treatment of cancer has thus evolved from broad chemotherapeutic approaches to therapies targeted towards some of these specific molecular abnormalities that drive tumor growth. To date, there are a few number of drugs approved by the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for NSCLC presenting specific molecular alterations (Table 1). However, an increasing number of agents targeting genetic alterations are being evaluated in clinical trials.

Full table
Therefore, in routine clinical practice, robust and accurate assessment of molecular alterations within tumors is mandatory to determine which patients are suitable for these targeted therapeutics. Molecular testing is performed using formalin fixed paraffin embedded (FFPE) tumor tissue obtained by biopsy or surgery. Adequate tumor samples (tissue or cytology) taken in a suitable form are clinically important for a complete pathological diagnosis including tumor typing and sub-typing, and analysis of predictive markers. Molecular testing guidelines and recommendations have been published recently (3,4).
However, it can be challenging to obtain sufficient tumor tissue for molecular testing (particularly when biopsy samples are small and/or a very few percentage of tumor cells are present or are prioritized for disease diagnosis). In addition, some invasive biopsy procedures may present with a health risk for some patients. In this context, up to 20–30% of NSCLC patients may be unable to provide a tumor sample suitable for molecular testing at diagnosis (5-7). Similarly, for patients progressing on treatment, a tumor rebiopsy is not always available or may not be of sufficient quality to allow molecular testing (8). Thus, analysis of circulating free tumor-derived DNA (ctDNA) has been proposed as an alternative or a complementary minimally invasive method for the detection of molecular alterations in NSCLC patients.
Cell-free nucleic acids and circulating tumor DNA
The presence of nucleic acids in the circulation was first reported by Mandel and Metais in 1948 (9). Circulating cell-free DNA (cfDNA) is a common constituent of blood samples, present at a very low concentration (5–10 ng/mL) in healthy individual (10,11). This basal level can be increased in inflammation (12), during pregnancy (13), and the concentration of cfDNA was first demonstrated to be increased in cancer patients in 1977 (14).
In NSCLC patients, the baseline concentration of ctDNA is correlated with tumor burden measured by CT scan (15-18), with tumor metabolism as assessed by PET scan (18-21) and to TNM stages (21,22). High concentrations of baseline ctDNA constitute a poor prognostic factor in progression-free survival (PFS) and overall survival (OS), independently of age, stage, nature of the treatment, histological subtype or smoking status (17,19,22,23). Thus, Yang et al. recently proposed to incorporate ctDNA analysis (blood-based liquid biopsy) in a modified TNMB staging system (24).
Circulating tumor DNA is a part of cfDNA coming from tumor cells. The process by which tumor DNA enters the bloodstream is not fully understood (25-27). The length of ctDNA is in the range 180–200 base pairs, suggesting that ctDNA is mainly released by apoptotic cells (28). Circulating tumor cells observed in NSCLC patients are usually in a quite low number, suggesting that these cells are probably not a major source of ctDNA. Moreover, it has been suggested that tumor cells may actively secrete DNA fragments via extracellular vesicles including exosomes (29-31).
CfDNA and ctDNA are also present in other biological fluids allowing, for instance, the detection of EGFR mutations in urine (32,33) and in spinal fluid (34-36), but this will not be detailed further in this review which will be focused on plasma-derived ctDNA.
Preanalytical steps
Blood collection and handling are key steps in order to optimize the chance to detect a molecular alteration. Plasma (not serum) should be used for cfDNA mutation analysis, preventing contamination of plasma samples by wild-type DNA released from circulating leukocytes during clotting (11,37). Common anticoagulants such as EDTA and citrate are both suitable for processing of blood samples for cfDNA analysis (38), but EDTA is by far the most used to date. Again, in order to prevent release of normal DNA from blood cells, it is recommended to process blood to plasma within 4 hours of draw (39). Alternatively, use of stabilization collection tubes containing fixatives, such as the Cell-Free DNA BCT tubes (Streck) (40,41) or the cell-free DNA collection tubes (Roche Diagnostics) (42) allow blood processing at a later time, up to 10 days after collection (43).
Plasma is obtained via centrifugation of the blood sample (1,200–2,000 g, 10 min, 25 °C). A second, high-speed spin must be performed before or after freeze/thaw (3,000−16,000 g, 3 min) in a microcentrifuge to generate clean samples for mutation analysis.
DNA extraction can then be performed using one of the numerous commercially available kits specifically designed to extract cfDNA from plasma.
Technical issues
The improvement in detection techniques has allowed to detect molecular alterations in ctDNA. In theory, all the molecular techniques allowing to detect a mutation can be used. But the fraction of ctDNA can be very low, therefore requiring highly sensitive techniques. Three main approaches are commonly used: allele-specific PCR (e.g., COBAS, Roche Diagnostics; Therascreen, Qiagen), digital PCR (dPCR) [including droplet digital PCR (ddPCR) and Beads, Emulsion, Amplification, and Magnetics (BEAMing)] and next generation sequencing (NGS). Several head-to-head comparisons have been performed (44-46), and detailed reviews have now been published (39,47,48). The main advantages and disadvantages of each technique are summarized in Table 2.

Full table
The first two approaches have in common that they are designed to detect specific alterations. This is convenient when the number of alterations which could be detected is limited (typically the EGFR T790M resistance mutation). But this is a limitation when a significant number of genes/alterations have to be analyzed at once. In such circumstances [ALK resistance mutations, Tumor Mutation Burden (TMB), …], NGS approaches are clearly required.
Clinical use of ctDNA testing
The clinical use of ctDNA analysis can be split in two categories:
- Detection of targetable molecular alteration (at diagnosis and/or at progression) is nowadays performed in routine practice. We will address the main issues related to these applications;
- Monitoring ctDNA over time could be useful for monitoring treatment efficiency and relapse in a relatively non-invasive way, but this is not yet used in routine practice. These potential future application of ctDNA testing in clinical practice will be discussed in the last part of this review.
EGFR: from activating mutations to resistance mutations
Many studies reported the detection of EGFR activating mutation in ctDNA of patients with NSCLC. Some of these studies have been included in meta-analyses (51-53). Altogether, these studies indicate that it is feasible to detect EGFR mutation in ctDNA, with in most cases a reasonable sensitivity (pooled sensitivity 62–75%) and a good specificity (pooled sensitivity 79–96%).
Prospective clinical trials have allowed to validate these findings with large series of patients (6,54-56). For instance, in the open-label IFUM study of Caucasian patients with EGFR mutation-positive NSCLC, a mutation status concordance of 94.3% [sensitivity 65.7%, specificity 99.8%, positive predictive value (PPV) 98.6%, negative predictive value (NPV) 93.8%] was observed between 652 matched tissue/cytology and plasma samples (6).
Finally, the multicentre, non-interventional, diagnostic ASSESS study investigated ctDNA for EGFR mutation testing in patients with advanced NSCLC in the real-world setting (57). Overall, the data obtained confirmed that ctDNA is a feasible sample type for real-world EGFR mutation testing, if robust and sensitive DNA extraction and mutation analysis methodologies are employed (57).
More sensitive techniques such as dPCR have shown higher sensitivity to detect EGFR mutations in NSCLC patients (58). In few cases, EGFR mutations detected at low levels in ctDNA were found to be subclonal in the tumor tissue (59). However, these are rare cases because EGFR mutations are almost always clonal in NSCLC patients (60).
Altogether, these data confirmed ctDNA as a powerful alternative sample for EGFR mutation analysis in patients with advanced NSCLC, particularly when no tissue sample is evaluable or available. In agreement with this conclusion, the EMA approved the use of ctDNA obtained from a blood (plasma) sample for EGFR mutation assessment before treatment with gefitinib in 2015. In June 2016, the FDA approved cobas EGFR Mutation Test v2 (Roche Molecular Systems, Inc.) using plasma specimens as a companion diagnostic test for the detection of exon 19 deletions or exon 21 mutations in the EGFR gene to identify patients with metastatic NSCLC eligible for treatment with erlotinib.
Most NSCLCs with activating EGFR alterations respond dramatically to tyrosine kinase inhibitors (TKIs). However, all these patients will ultimately relapse. The most frequent alteration associated with TKI resistance is the EGFR-T790M gatekeeper mutation (61-64). Third generation EGFR TKIs have been developed to irreversibly inhibit mutant EGFR, including the EGFR T790M variant. Osimertinib has shown to be very efficient in T790M-positive patients (65), and was thus first approved in this context. In the clinical setting, it is therefore mandatory to test EGFR-mutated patients treated with EGFR inhibitors for the presence of the T790M mutation at relapse. The European approval of osimertinib indicates that this test can be performed using either tumour DNA derived from a tissue sample or ctDNA obtained from a plasma sample. In this context, liquid biopsies using ctDNA have distinct advantages over traditional biopsy methods (listed in Table 3). If this mutation is found, the patient can be treated with osimertinib. Indeed EGFR-mutant NSCLC patients with T790M mutation detected by ctDNA benefit treatment with osimertinib (69). However, if the ctDNA test is negative, it is advisable to follow-up with a tissue test wherever possible due to the potential for false negative results using a plasma-based test (39,70).
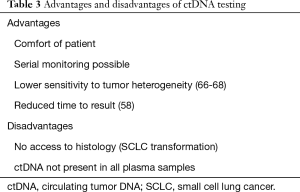
Full table
At the molecular level, the T790M mutation is a single nucleotide change (c.2369C>T). In this context, techniques that focus on specific alterations in order to be as sensitive as possible might be adequate for this analysis. dPCR approaches are the most sensitive techniques in this context (44), even if the COBAS assay turned out to be the most robust approach in the first round of external quality assessments performed in France (71).
T790M-positive patients treated with osimertinib finally relapse and acquire new molecular alterations. Novel mutations of the EGFR gene associated with resistance to osimertinib including the C797S mutation (72-75) and others (76) have been described. A variety of additional alterations including KRAS mutation (74), BRAF V600 mutation, HER2 amplification and MET amplification have also detected in tumors and/or ctDNA collected at progression after osimertinib (77-80). When novel treatments targeting these alterations will be available/approved, it will be most convenient to look for all these alterations in ctDNA by using NGS approaches.
Osimertinib has recently been shown to be also very efficient in first line treatment of EGFR mutated patients (81,82). At progression, patients do not develop the T790M mutation (80). Therefore, if the use of osimertinib in frontline becomes a standard of care, the T790M testing in ctDNA will not be required anymore.
On the other hand, patients progressing on first line osimertinib have been shown to develop molecular mechanisms of resistance that are similar to those described in second line of treatment. Some treatment naive patients have been enrolled in the AURA trial (80). Plasma samples were collected at progression on osimertinib and several potential resistance mechanisms were reported (80). The EGFR C797S mutation was detected in 2 patients. Interestingly, cells lines presenting the C797S mutation (but not the T790M) in addition to an activating mutation were sensitive to first generation (gefitinib) and second generation (afatinib) EGFR TKI inhibitors (83), suggesting that these inhibitors could be efficient as second line treatment in this context. Other alterations including amplification of MET, EGFR and KRAS, mutation of MEK1, KRAS, PIK3CA, HER2 and JAK2 have been described in patients progressing on osimertinib treatment (80). Gene fusion have also been reported as mechanisms of resistance to osimertinib (84,85). Recent reports indicated that some of these alterations can be successfully targeted (84,86-88). Therefore, the analysis of ctDNA collected at progression in patients treated with osimertinib front-line will be most useful to guide effective second-line therapy.
ALK: from gene rearrangements to resistance mutation
In routine practice, the detection of ALK rearrangements is usually performed by immunohistochemistry and fluorescent in-situ hybridization (FISH) on tissue specimens (89-91). NGS-based gene capture approaches using DNA from FFPE samples have been described (92). Using a similar approach, McCoach et al. were able to detect ALK fusions in ctDNA (93).
RNA-based strategies (RT-PCR and RNA sequencing) have also been demonstrated to detect ALK rearrangements in tissue samples with a high sensitivity and specificity, and allowing to identify ALK fusion variants (94-97). RT-PCR has also been used to detect fusion transcripts in circulating RNA (98,99), but these approaches are not yet used in routine clinical practice.
Secondary ALK mutations that confer acquired resistance to crizotinib have been described (100,101). These alterations do not appear to prevent efficacy of second generation ALK inhibitors such as ceritinib (102). Other acquired mutations have been described upon treatment with second line ALK inhibitors (103). These mutations can be detected both in tumor tissues and in ctDNA (101). Since many different mutations have been described, NGS approaches are the most appropriate techniques in this context. However, there is currently insufficient evidence to support the use of testing ALK mutational status for lung adenocarcinoma patients with sensitizing ALK translocation who have progressed after treatment with an ALK-targeted TKI (3).
BRAF gene
The BRAF V600E mutation is frequent in metastatic melanoma, and a number of studies reported the clinical use of ctDNA testing in this context (10,104-106). This mutation is much less frequent in NSCLC (approximately 1% of cases). Recent clinical trials have demonstrated a significant effect of the combination of a BRAF inhibitor combined to a MEK inhibitor (107). Several reports demonstrated a clinical use of ctDNA equivalent to that described for EGFR (108,109).
Mutation load and other genetic/epigenetic alterations
Several clinical trials recently demonstrated that a high TMB is associated with improved efficacy of PD-1/PD-L1 inhibitors (110,111). Interestingly, Gandara and colleagues have recently demonstrated that it is possible to use ctDNA to determine TMB (112). They used plasma samples collected in two large randomized trials (OAK and POLAR), evaluating atezolizumab (anti-PD-L1) in second-line and higher. The TMB score was determined by identifying all base substitutions present at an allele frequency of ≥0.5% across the coding region of 394 genes (1.1 Mb). They demonstrated a relationship between clinical outcomes and the ctDNA TMB (112). Further studies are necessary to establish the conditions of using this test in routine clinical practice, in particular the cut off value that should be used.
Many different genetic or epigenetic alterations, such as of gene methylation (113-115) or detection of microsatellite alterations (116,117) can be detected using ctDNA. But detection of these modifications has no clinical application at present.
ctDNA and minimal residual disease (MRD)
At early and locally advanced stages, the reference treatment of NSCLC remains surgical resection, combined with adjuvant chemotherapy or chemoradiotherapy. Despite therapeutic developments in this area, cancer recurrence remains the leading cause of postoperative NSCLC mortality, with significant relapse rates, while the OS benefit of adjuvant chemotherapy remains low (118). In a meta-analysis of five clinical trials of 4,584 patients, Pignon et al. showed that adjuvant chemotherapy had a deleterious effect on OS in patients with stage IA, and possibly stage IB NSCLC, while the recurrence rate was up to 36% in these patients (119). This issue has generated a great interest in the development of markers predictive of postoperative recurrence, to strengthen adjuvant therapies and postoperative follow-up in patients at risk of recurrence, while limiting exposure to cytotoxic agents and ionizing radiations in low-risk patients.
As the detection of ctDNA in plasma indicates the presence of residual tumor tissue, it has been proposed to use this biomarker to assess the MRD after surgical resection in locally operable advanced NSCLCs. Chaudhuri et al. showed that post-surgical detection of ctDNA was associated in 100% of cases (20/20 patients) with recurrence of the disease (21). In addition, detection of ctDNA preceded radiological detection of progression in 72% of patients, with a median of 5.2 months. The detectability of ctDNA in this study was an independent prognostic factor: patients with detectable ctDNA on a sample collected less than 4 months after surgery had a very significantly lower PFS and OS than those for whom ctDNA was undetectable (36-month PFS =0% vs. 93%, respectively). Nevertheless, at the end of the study, 6% of patients with undetectable ctDNA also had a recurrence. Similar results have been observed in other studies with NSCLC patients (6,14,15,18,120,121).
While the use of ctDNA for post-surgical MRD evaluation of NSCLC appears promising, the interpretation of an undetectable ctDNA remains difficult because of the limited detection sensitivity of molecular biology tests, particularly after the surgical reduction of the tumor burden. Prospective studies are required to determine whether the use of adjuvant therapy based on the detection of ctDNA would improve clinical outcomes. Following surgery, patients will be tested on plasma. If the test is negative, the patients will be followed until progression, and then treated with adjuvant therapy. If the initial plasma test is positive, patients will be randomized. In the control arm, the patients will be followed until progression, and then treated, as patients with a negative plasma test. In the experimental arm, the patients will be directly treated with adjuvant therapy (Figure 1).
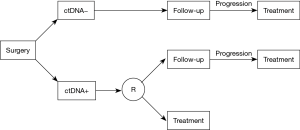
ctDNA monitoring and early assessment of response
ctDNA testing is also an appealing way of monitoring the activity of systemic treatments in the metastatic stage. Numerous studies evaluated the correlation between therapeutic response and longitudinal quantitative changes in plasma ctDNA. The first ctDNA monitoring applications for therapeutic follow-up implicated cohorts of patients treated with EGFR TKI for advanced NSCLC. The existence of a mutation of the EGFR gene, common to all these patients, allowed quantification of ctDNA by quantitative targeted techniques, such as dPCR.
For instance, Mok et al. showed that in 66 patients treated with erlotinib plus gemcitabine for an EGFR-mutated NSCLC and whose activating mutation was detectable at baseline, the response rate of patients whose ctDNA had become undetectable at 12 weeks was greater to that of patients whose ctDNA remained detectable at 12 weeks (83% vs. 67%) (122). The undetectability of ctDNA at 12 weeks was also associated with a significant benefit in PFS and OS [hazard ratio (HR) =0.38, P=0.0083 and HR =0.38, P=0.0831, respectively] (122).
More recently, Taus et al. observed changes in ctDNA in patients treated with TKI or chemotherapy for advanced EGFR-mutated NSCLC, whose EGFR-activating mutation was detectable in dPCR at baseline (123). A decrease in ctDNA was observed in 13 of 14 evaluable cases (93%), 38 days before the radiological response in median (20 to 69 days). Conversely, in 17 of 19 evaluable cases (89%), an increase in ctDNA was found 80 days in median (12 to 292 days) before radiological progression assessed by CT scan. Patients whose circulating EGFR mutation became undetectable during follow-up had a significantly better PFS than those for whom ctDNA was still detectable (median: 295 vs. 55 days, respectively).
The recent development of immune checkpoint immunotherapies has generated a particular interest in early identification of therapeutic response by longitudinal analysis of ctDNA concentrations. Indeed, the response to immunotherapies may be difficult to assess on imaging, because it can be characterized by an increase in the apparent tumor volume or the appearance of new lesions, due to the leukocyte infiltration of the tumor that it induces. Despite the development of radiological criteria adapted to immunotherapies, the identification of non-responders remains late (124). One of the main difficulties associated with ctDNA monitoring in these patients is that they do not carry consensual genetic alteration: for this reason, most of the studies evaluating the ctDNA interest in this application have quantified ctDNA in NGS, by measuring the variant allelic frequency (VAF) of mutations identified by screening of a large panel of genes.
Giroux Leprieur et al. recently demonstrated, on a cohort of 15 patients treated with nivolumab and with detectable somatic alteration at baseline in NGS, that the absence of a significant increase in VAF at 2 months (defined as an increase of more than 9% relative to baseline) predicted a disease control of at least 6 months with sensitivity of 71% and specificity of 100% (125). Moreover, the absence of ctDNA increase was associated with a significantly higher PFS and OS (median: 0.7 vs. 12.0 and 2.1 months vs. not-reached, respectively) (125).
In another study, Goldberg et al. defined a “ctDNA response” as a decrease in VAF greater than 50% of the baseline, and confirmed on a second sample (126). This study, carried out on 28 patients treated by anti-PD-1 or anti-PD-L1 immunotherapy and carrying a somatic detectable alteration at baseline in NGS, showed that patients presenting a “ctDNA response” during follow-up presented a longer duration of treatment, PFS and OS than those who did not (median: 206 vs. 69 days; HR =0.17, 95% CI: 0.05–1.02 and HR =0.13, 95% CI: 0.03–0.51, respectively). On this cohort, the “ctDNA response” was obtained 42.5 days before the radiological confirmation of the response on CT scan, in median (126).
Finally, Raja et al. conducted an NGS analysis of 28 durvalumab-treated patients with somatic detectable alteration at baseline (127). The changes in the VAF of this mutation were correlated with the therapeutic response, with a mean decrease of 2.7% in responder patients compared to a mean increase of 1.7% in non-responder patients. The authors also showed that a decrease in VAF at 6 weeks of treatment was associated with a benefit in PFS and OS (median: 1.45 vs. 13.7 months and 9.07 vs. 28.13 months). The decrease in ctDNA preceded from 1 to 12 months the radiological confirmation of the therapeutic response in 70% of patients (127).
Some studies have also demonstrated the existence of a peak concentration of ctDNA very early after the start of treatment, probably related to the massive release of tumor DNA by treatment-induced cell lysis (128,129). Whether this very early increase of ctDNA is associated with a better outcome of patients remains to be determine in a large prospective study.
Overall, all the studies evaluating the relevance of kinetic analysis of ctDNA for predicting response of NSCLC to systemic therapies tend to show that the reduction of ctDNA at the beginning of treatment makes it possible to predict the therapeutic response earlier than imaging and is associated with a more favorable prognosis in PFS and OS. A significant increase in ctDNA concentration during follow-up also seems to predict disease progression earlier than radiological monitoring. These data have not yet been applied in the management of patients in clinical practice. Thus, clinical trials are required. A design for such trials is presented on Figure 2. In the reference arm, patients will be treated with A, then with B following radiological or clinical progression. In the experimental arm, early ctDNA analysis (after for instance 2 or 3 weeks of treatment), will allow to identify a change in ctDNA concentration as compared to the pre-treatment assay performed. In case of significant decrease, suggesting that the patient is responding, treatment A will be continued until radiological or clinical progression. If there is no decrease in ctDNA concentration, suggesting that the patient is not responding to treatment A, the treatment will be changed to B.
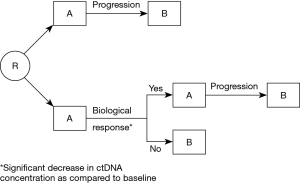
Such use of ctDNA kinetics will be limited by the need to identify a molecular alteration in the patients’ tumor, which is not always the case, even using broad NGS approaches. Furthermore, some patients have no ctDNA detectable at baseline.
It might be also necessary to use techniques allowing an absolute quantification of ctDNA. NGS allows only a relative quantification of the mutated copies, as compared to the wild-type alleles. But multiple physiopathological or preanalytical factors are likely to induce an increased release of non-tumor DNA into the plasma, including inflammation, stimulation of antitumor immunity, lysis of the healthy parenchyma during tumor progression, and in vitro leukocyte lysis related to a delay in sample processing or a traumatic puncture (130). dPCR allows an absolute quantification of the mutated copies concentration in the sample, independently of the “background noise” induced by the presence of non-tumor DNA. However, it remains a targeted technique since each test allows the quantification of only a limited number of previously determined mutations on the tumor tissue. Demuth et al. recently proposed a test protocol combining a broad screening of ctDNA at baseline by NGS to identify one or more somatic mutations, and an absolute quantification of these mutations on circulating DNA during follow-up, by custom dPCR analyzes for each patient according to the alterations found in NGS (131).
Finally, the lack of consensual evaluation criteria for longitudinal variations of ctDNA is one of the major limitations to the use of this biomarker. Indeed, many studies base their results on the notion of detectability of somatic mutations on circulating DNA. However, this notion is particularly dependent on the pre-analytical and analytical processes used: in the absence of a common methodology, the results of these different studies can therefore hardly be compared and reproduced. For the analysis of quantitative variations of ctDNA during follow-up the accuracy of the method used should be considered, and it depends on the number of mutated copies in the sample (for dPCR) (132) or on the allelic frequency (for NGS) (133). In a recent study on the monitoring of metastatic cutaneous melanoma treated with anti-PD-1 immunotherapy, we recently proposed interpretation criteria based on the point-to-point statistical comparison of ctDNA concentrations measured during follow-up. We defined the biological response (bR) as a significant decrease in ctDNA compared to the baseline measurement, given the inaccuracy of the two measurements. Biological progression (bP) was defined as a significant increase in ctDNA compared to the nadir measurement. In an evaluation of 22 patients, bP was predictive of progression on average 79 days before radiological progression, with 100% sensitivity and 100% specificity, while bR predicted the therapeutic response to average 115 days before the objective radiological response with a sensitivity of 100% and a specificity of 50%. We also showed that the absence of bR at the 2nd week of treatment, in 10/22 patients, was associated with a lack of clinical benefit, with a 0% PFS rate at 4 months (105). The development and evaluation of similar criteria incorporating the inaccuracy of the ctDNA measurement for the interpretation of its variations could improve the predictive value of this biomarker for the therapeutic monitoring of NSCLC.
Conclusions
ctDNA is already used in routine practice for the detection of molecular alterations, allowing to guide therapy. When new drugs targeting other genes will be approved, it will be easy to use this source of tumor DNA to set up these new assays.
More work is required to determine whether a therapeutic strategy guided by ctDNA analysis can improve PFS, OS and patients’ quality of life as compared to radiological/clinical monitoring.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. [Crossref] [PubMed]
- Shames DS, Wistuba II. The evolving genomic classification of lung cancer. J Pathol 2014;232:121-33. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Akerley W, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. J Natl Compr Canc Netw 2016;14:255-64. [Crossref] [PubMed]
- NLCA annual report 2015. Available online: https://www.rcplondon.ac.uk/projects/outputs/nlca-annual-report-2015
- Douillard JY, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer 2014;110:55-62. [Crossref] [PubMed]
- Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol 2015;26:1415-21. [Crossref] [PubMed]
- Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: a prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014;86:170-3. [Crossref] [PubMed]
- Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Herbreteau G, Vallee A, Knol AC, et al. Circulating tumour DNA: analytical aspects and clinical applications for metastatic melanoma patients. Ann Biol Clin (Paris) 2017;75:619-30. [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [Crossref] [PubMed]
- Leon SA, Ehrlich GE, Shapiro B, et al. Free DNA in the serum of rheumatoid arthritis patients. J Rheumatol 1977;4:139-43. [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Zhu YJ, Zhang HB, Liu YH, et al. Quantitative cell-free circulating EGFR mutation concentration is correlated with tumor burden in advanced NSCLC patients. Lung Cancer 2017;109:124-7. [Crossref] [PubMed]
- Tsui DWY, Murtaza M, Wong ASC, et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO Mol Med 2018;10. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Winther-Larsen A, Demuth C, Fledelius J, et al. Correlation between circulating mutant DNA and metabolic tumour burden in advanced non-small cell lung cancer patients. Br J Cancer 2017;117:704-9. [Crossref] [PubMed]
- Nygaard AD, Holdgaard PC, Spindler KL, et al. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 2014;110:363-8. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Hyun MH, Sung JS, Kang EJ, et al. Quantification of circulating cell-free DNA to predict patient survival in non-small-cell lung cancer. Oncotarget 2017;8:94417-30. [PubMed]
- Lee Y, Park S, Kim WS, et al. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stage EGFR-mutated non-small cell lung cancer. Thorac Cancer 2018;9:1104-10. [Crossref] [PubMed]
- Yang M, Forbes ME, Bitting RL, et al. Incorporating blood-based liquid biopsy information into cancer staging: time for a TNMB system? Ann Oncol 2018;29:311-23. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139-42. [Crossref] [PubMed]
- Stroun M, Maurice P, Vasioukhin V, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci 2000;906:161-8. [Crossref] [PubMed]
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347-76. [Crossref] [PubMed]
- Stroun M, Anker P, Lyautey J, et al. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 1987;23:707-12. [Crossref] [PubMed]
- Anker P, Stroun M, Maurice PA. Spontaneous extracellular synthesis of DNA released by human blood lymphocytes. Cancer Res 1976;36:2832-9. [PubMed]
- Castellanos-Rizaldos E, Grimm DG, Tadigotla V, et al. Exosome-Based Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin Cancer Res 2018;24:2944-50. [Crossref] [PubMed]
- Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol 2018;29:700-6. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Tchekmedyian N, Mudad R, Blanco FF, et al. Longitudinal monitoring of ctDNA EGFR mutation burden from urine correlates with patient response to EGFR TKIs: A case series. Lung Cancer 2017;108:22-8. [Crossref] [PubMed]
- Theoleyre S, Masson I, Herbreteau G, et al. Treatment of a NSCLC patient with osimertinib based on the detection of the EGFR T790M resistance mutation in cerebrospinal fluid. Lung Cancer 2017;114:111-2. [Crossref] [PubMed]
- Yang H, Cai L, Zhang Y, et al. Sensitive detection of EGFR mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn 2014;16:558-63. [Crossref] [PubMed]
- Zhao J, Ye X, Xu Y, et al. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol 2016;78:1305-10. [Crossref] [PubMed]
- Vallee A, Marcq M, Bizieux A, et al. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 2013;82:373-4. [Crossref] [PubMed]
- Lam NY, Rainer TH, Chiu RW, et al. EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin Chem 2004;50:256-7. [Crossref] [PubMed]
- Normanno N, Denis MG, Thress KS, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017;8:12501-16. [Crossref] [PubMed]
- Norton SE, Lechner JM, Williams T, et al. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem 2013;46:1561-5. [Crossref] [PubMed]
- Norton SE, Luna KK, Lechner JM, et al. A new blood collection device minimizes cellular DNA release during sample storage and shipping when compared to a standard device. J Clin Lab Anal 2013;27:305-11. [Crossref] [PubMed]
- Alidousty C, Brandes D, Heydt C, et al. Comparison of Blood Collection Tubes from Three Different Manufacturers for the Collection of Cell-Free DNA for Liquid Biopsy Mutation Testing. J Mol Diagn 2017;19:801-4. [Crossref] [PubMed]
- Denis MG, Knol AC, Theoleyre S, et al. Efficient Detection of BRAF Mutation in Plasma of Patients after Long-term Storage of Blood in Cell-Free DNA Blood Collection Tubes. Clin Chem 2015;61:886-8. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Pasquale R, Fenizia F, Esposito Abate R, et al. Assessment of high-sensitive methods for the detection of EGFR mutations in circulating free tumor DNA from NSCLC patients. Pharmacogenomics 2015;16:1135-48. [Crossref] [PubMed]
- Keppens C, Palma JF, Das PM, et al. Detection of EGFR Variants in Plasma: A Multilaboratory Comparison of a Real-Time PCR EGFR Mutation Test in Europe. J Mol Diagn 2018;20:483-94. [Crossref] [PubMed]
- Liang Z, Cheng Y, Chen Y, et al. EGFR T790M ctDNA testing platforms and their role as companion diagnostics: Correlation with clinical outcomes to EGFR-TKIs. Cancer Lett 2017;403:186-94. [Crossref] [PubMed]
- Moding EJ, Diehn M, Wakelee HA. Circulating tumor DNA testing in advanced non-small cell lung cancer. Lung Cancer 2018;119:42-7. [Crossref] [PubMed]
- Garcia J, Dusserre E, Cheynet V, et al. Evaluation of pre-analytical conditions and comparison of the performance of several digital PCR assays for the detection of major EGFR mutations in circulating DNA from non-small cell lung cancers: the CIRCAN_0 study. Oncotarget 2017;8:87980-96. [Crossref] [PubMed]
- Decraene C, Silveira AB, Bidard FC, et al. Multiple Hotspot Mutations Scanning by Single Droplet Digital PCR. Clin Chem 2018;64:317-28. [Crossref] [PubMed]
- Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev 2015;24:206-12. [Crossref] [PubMed]
- Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269. [Crossref] [PubMed]
- Qian X, Liu J, Sun Y, et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016;7:29154-65. [Crossref] [PubMed]
- Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. [Crossref] [PubMed]
- Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer 2017;116:175-85. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Rachiglio AM, Esposito Abate R, Sacco A, et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 2016;7:66595-605. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Denis MG, Vallee A, Theoleyre S. EGFR T790M resistance mutation in non small-cell lung carcinoma. Clin Chim Acta 2015;444:81-5. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in Pretreated T790M-Positive Advanced Non-Small-Cell Lung Cancer: AURA Study Phase II Extension Component. J Clin Oncol 2017;35:1288-96. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017;28:784-90. [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Denis M, Vallée A, Charpentier S, et al. Nationwide external quality assessment (EQA) of EGFR testing in circulating tumor DNA: The French experience. Ann Oncol 2017;28:v22-42. [Crossref]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Wang Z, Yang JJ, Huang J, et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J Thorac Oncol 2017;12:1723-7. [Crossref] [PubMed]
- Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin Cancer Res 2016;22:4837-47. [Crossref] [PubMed]
- Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018;29:1049-55. [Crossref] [PubMed]
- Chen K, Zhou F, Shen W, et al. Novel Mutations on EGFR Leu792 Potentially Correlate to Acquired Resistance to Osimertinib in Advanced NSCLC. J Thorac Oncol 2017;12:e65-8. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Lin CC, Shih JY, Yu CJ, et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med 2018;6:107-16. [Crossref] [PubMed]
- Le X, Puri S, Negrao MV, et al. Landscape of EGFR -dependent and -independent resistance mechanisms to osimertinib and continuation therapy post-progression in EGFR-mutant NSCLC. Clin Cancer Res 2018;24:6195-203. [PubMed]
- Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:841-9. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Reungwetwattana T, Nakagawa K, Cho BC, et al. CNS Response to Osimertinib Versus Standard Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Niederst MJ, Hu H, Mulvey HE, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res 2015;21:3924-33. [Crossref] [PubMed]
- Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529-39. [Crossref] [PubMed]
- Zeng L, Yang N, Zhang Y. GOPC-ROS1 Rearrangement as an Acquired Resistance Mechanism to Osimertinib and Responding to Crizotinib Combined Treatments in Lung Adenocarcinoma. J Thorac Oncol 2018;13:e114-6. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. [Crossref] [PubMed]
- Robichaux JP, Elamin YY, Tan Z, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 2018;24:638-46. [Crossref] [PubMed]
- Fassunke J, Muller F, Keul M, et al. Overcoming EGFR(G724S)-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun 2018;9:4655. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823-59. [Crossref] [PubMed]
- Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:322-8. [Crossref] [PubMed]
- Lantuejoul S, Rouquette I, Blons H, et al. French multicentric validation of ALK rearrangement diagnostic in 547 lung adenocarcinomas. Eur Respir J 2015;46:207-18. [Crossref] [PubMed]
- Jang JS, Wang X, Vedell PT, et al. Custom Gene Capture and Next-Generation Sequencing to Resolve Discordant ALK Status by FISH and IHC in Lung Adenocarcinoma. J Thorac Oncol 2016;11:1891-900. [Crossref] [PubMed]
- McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:2758-70. [Crossref] [PubMed]
- McLeer-Florin A, Duruisseaux M, Pinsolle J, et al. ALK fusion variants detection by targeted RNA-next generation sequencing and clinical responses to crizotinib in ALK-positive non-small cell lung cancer. Lung Cancer 2018;116:15-24. [Crossref] [PubMed]
- Dacic S, Villaruz LC, Abberbock S, et al. ALK FISH patterns and the detection of ALK fusions by next generation sequencing in lung adenocarcinoma. Oncotarget 2016;7:82943-52. [Crossref] [PubMed]
- Letovanec I, Finn S, Zygoura P, et al. Evaluation of NGS and RT-PCR Methods for ALK Rearrangement in European NSCLC Patients: Results from the European Thoracic Oncology Platform Lungscape Project. J Thorac Oncol 2018;13:413-25. [Crossref] [PubMed]
- Vaughn CP, Costa JL, Feilotter HE, et al. Simultaneous detection of lung fusions using a multiplex RT-PCR next generation sequencing-based approach: a multi-institutional research study. BMC Cancer 2018;18:828. [Crossref] [PubMed]
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Tong Y, Zhao Z, Liu B, et al. 5'/ 3' imbalance strategy to detect ALK fusion genes in circulating tumor RNA from patients with non-small cell lung cancer. J Exp Clin Cancer Res 2018;37:68. [Crossref] [PubMed]
- Duchemann B, Friboulet L, Besse B. Therapeutic management of ALK+ nonsmall cell lung cancer patients. Eur Respir J 2015;46:230-42. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Denis MG, Khammari A, Vallee A, et al. Detection of BRAF mutations in the plasma of melanoma patients as an early marker of treatment efficiency. J Clin Oncol 2014;32:abstr 9069.
- Herbreteau G, Vallee A, Knol AC, et al. Quantitative monitoring of circulating tumor DNA predicts response of cutaneous metastatic melanoma to anti-PD1 immunotherapy. Oncotarget 2018;9:25265-76. [Crossref] [PubMed]
- Quereux G, Herbreteau G, Knol AC, et al. Efficient treatment of a metastatic melanoma patient with a combination of BRAF and MEK inhibitors based on circulating tumor DNA analysis: a case report. BMC Res Notes 2017;10:320. [Crossref] [PubMed]
- Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017;18:1307-16. [Crossref] [PubMed]
- Guibert N, Pradines A, Casanova A, et al. Detection and Monitoring of the BRAF Mutation in Circulating Tumor Cells and Circulating Tumor DNA in BRAF-Mutated Lung Adenocarcinoma. J Thorac Oncol 2016;11:e109-12. [Crossref] [PubMed]
- Yang Y, Shen X, Li R, et al. The detection and significance of EGFR and BRAF in cell-free DNA of peripheral blood in NSCLC. Oncotarget 2017;8:49773-82. [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Legrand FA, Gandara DR, Mariathasan S, et al. Association of high tissue TMB and atezolizumab efficacy across multiple tumor types. J Clin Oncol 2018;36:12000. [Crossref]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin Chem Lab Med 2016;54:1385-93. [Crossref] [PubMed]
- Gainetdinov IV, Kapitskaya KY, Rykova EY, et al. Hypomethylation of human-specific family of LINE-1 retrotransposons in circulating DNA of lung cancer patients. Lung Cancer 2016;99:127-30. [Crossref] [PubMed]
- Wang BH, Li YY, Han JZ, et al. Gene methylation as a powerful biomarker for detection and screening of non-small cell lung cancer in blood. Oncotarget 2017;8:31692-704. [PubMed]
- Sanchez-Cespedes M, Monzo M, Rosell R, et al. Detection of chromosome 3p alterations in serum DNA of non-small-cell lung cancer patients. Ann Oncol 1998;9:113-6. [Crossref] [PubMed]
- Bruhn N, Beinert T, Oehm C, et al. Detection of microsatellite alterations in the DNA isolated from tumor cells and from plasma DNA of patients with lung cancer. Ann N Y Acad Sci 2000;906:72-82. [Crossref] [PubMed]
- Isbell JM, Jones DR, Li BT. Circulating tumor DNA: A promising biomarker to guide postoperative treatment and surveillance of non-small cell lung cancer. J Thorac Cardiovasc Surg 2018;155:2628-31. [Crossref] [PubMed]
- Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. [Crossref] [PubMed]
- Hu W, Yang Y, Zhang L, et al. Post surgery circulating free tumor DNA is a predictive biomarker for relapse of lung cancer. Cancer Med 2017;6:962-74. [Crossref] [PubMed]
- Liang H, Huang J, Wang B, et al. The role of liquid biopsy in predicting post-operative recurrence of non-small cell lung cancer. J Thorac Dis 2018;10:S838-45. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Taus Á, Camacho L, Rocha P, et al. Dynamics of EGFR Mutation Load in Plasma for Prediction of Treatment Response and Disease Progression in Patients With EGFR-Mutant Lung Adenocarcinoma. Clin Lung Cancer 2018;19:387-394.e2. [Crossref] [PubMed]
- Wolchok JD, Hoos A, O'Day S, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res 2009;15:7412-20. [Crossref] [PubMed]
- Giroux Leprieur E, Herbretau G, Dumenil C, et al. Circulating tumor DNA evaluated by Next-Generation Sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. Oncoimmunology 2018;7:e1424675. [Crossref] [PubMed]
- Goldberg SB, Narayan A, Kole AJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res 2018;24:1872-80. [Crossref] [PubMed]
- Raja R, Kuziora M, Brohawn PZ, et al. Early Reduction in ctDNA Predicts Survival in Patients with Lung and Bladder Cancer Treated with Durvalumab. Clin Cancer Res 2018;24:6212-22. [Crossref] [PubMed]
- Vallee A, Audigier-Valette C, Herbreteau G, et al. Rapid clearance of circulating tumor DNA during treatment with AZD9291 of a lung cancer patient presenting the resistance EGFR T790M mutation. Lung Cancer 2016;91:73-4. [Crossref] [PubMed]
- Kato K, Uchida J, Kukita Y, et al. Transient appearance of circulating tumor DNA associated with de novo treatment. Sci Rep 2016;6:38639. [Crossref] [PubMed]
- Lee TH, Montalvo L, Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001;41:276-82. [Crossref] [PubMed]
- Demuth C, Winther-Larsen A, Madsen AT, et al. A method for treatment monitoring using circulating tumour DNA in cancer patients without targetable mutations. Oncotarget 2018;9:31066-76. [Crossref] [PubMed]
- Majumdar N, Wessel T, Marks J. Digital PCR modeling for maximal sensitivity, dynamic range and measurement precision. PLoS One 2015;10:e0118833. [Crossref] [PubMed]
- Liu D, Zhou H, Shi D, et al. Quality Control of Next-generation Sequencing-based In vitro Diagnostic Test for Onco-relevant Mutations Using Multiplex Reference Materials in Plasma. J Cancer 2018;9:1680-8. [Crossref] [PubMed]


