
A pilot study of blood microRNAs and lung function in young healthy adults with fine particulate matter exposure
Introduction
Particulate matter (PM) has been brought to public attention. Fine particulate matter (PM2.5) are the PM with a diameter of 2.5 µm or less. According to the statement presented by Beijing Municipal Environmental Protection Bureau (1,2), PM2.5 is the main pollutants in Beijing, China. Once inhaled, PM2.5 deposits in lung tissues inducing airway and systematic inflammation (3). PM2.5 has been acknowledged as an essential risk factor of pulmonary diseases such as chronic obstructive pulmonary disease (COPD) (4). However, the mechanism is still unclear.
MicroRNAs (miRNAs) are a class of endogenous small noncoding RNA as negative regulators of gene expression via complementary base pairing to the 3’-untranslatedregion (3’-UTR) of their target mRNA (5). miRNAs have been recognized as ideal research tools because of highly conserved mechanism in eukaryotes. Different miRNAs have been shown to be associated with COPD, participating in physiological functions and inflammatory regulation (6-8). Recently, miRNAs are suggested to be potential indicators of environmental exposure such as miR-9, miR-10b and miR-21 in human (9). Recently, we reported that 10 differential miRNAs (miR-146a, -146b, -139, -129, -340, -691, -181a, -155, -21-3p, and -21-5p) were up-regulated in PM2.5 exposed mice (10). It suggested that miRNAs might play the role in the toxicological pathways of PM2.5 exposure. However, there were rarely studies looking for miRNAs of both COPD and PM2.5 exposure.
We hypothesized that it might be more targeted to study the relationship between COPD related miRNAs and PM2.5 exposure. Thus, we designed this pilot study. Firstly, we identified differentially expressed blood miRNAs contrasting COPD vs. non-COPD individuals. Next, we designed an independent time-series study to observe the expression of these significant miRNAs in healthy adults before and after PM2.5 exposure. As blood sample other than lung tissue is much easier and more ethical to obtain in healthy adults, this pilot study aimed to find some blood for further analyzing the mechanism of PM2.5.
Methods
Study design and participants
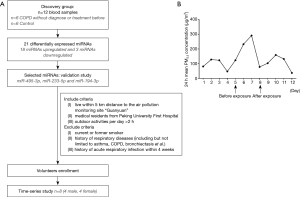
To find disease specific miRNAs, 12 blood samples from 6 COPD patients and 6 control participants were chosen from clinical trial (registered at www.ClinicalTrials.gov: NCT02037828). COPD patients were defined as forced expiratory volume in one second (FEV1)/forced vital capacity (FVC) <70% (post-bronchodilation) without diagnose of COPD or related treatment before. Control participants were selected with matched age, sex, body mass index (BMI) and smoking history.
To observe the expression of these differentially expressed miRNAs before and after exposure, a time series study was designed nested within “low-high-low” pollution levels, according to the air polluted weather forecasting and warning. Eight healthy volunteers (4 males and 4 females) from Peking University First Hospital were enrolled with informed consent. Volunteers were measured for lung function every day from Nov. 28 to Dec. 09 in 2016 and asked not to wear masks during the study. Blood samples were collected in Dec. 02 and Dec. 05, defined as “before exposure” and “after exposure” respectively.
The study was in compliance with the Declaration of Helsinki and was approved by Peking University Institutional Review Board Office (IRB00001052-13043) and Peking University First Hospital Institutional Review Board Office (2016-[1254]). The include/exclude criteria and flow chart of the study was shown in Figure 1A, and the demographic and characteristics of all participants were summarized in Table 1.
Full table
Definition of before or after exposure
There are 35 monitoring stations in Beijing and each station reports real-time concentration of PM2.5 every hour. 24-hour mean concentrations of PM2.5 were recorded according to the information platform from Beijing Municipal Environmental Monitoring Center (Bureau). In validation study, volunteers were living and working within the range of “Guanyuan” motoring station, so PM2.5 concentrations of “Guanyuan” were obtained (Figure 1B).
In our previous study, FVC and FEV1 showed negative dose-response associations with PM2.5 concentrations at lag0 (11), while blood biomarkers such as neutrophil count, fibrinogen had positive associations with PM2.5 concentrations at lag1–3 (12,13). These results indicated that there were delayed reactions of inhaling PM2.5 occurring in lungs first at lag0, and then in blood at lag1. In this study, blood samples were collected at the day with lag-1 concentration <50 µg/m3 of PM2.5 as “before exposure” and the day with lag-1 concentration >200 µg/m3 of PM2.5 as “after exposure”.
Human peripheral blood mononuclear cells (PBMCs) and RNA isolation
Totally, 4 mL blood samples were obtained using ethylenediaminetetraacetic acid (EDTA)-anticoagulation tube. PBMCs were extracted using Lymphoprep™ (Alere Technologies, P.O. Box 6863 Rodeløkka, N-0504 Oslo, Norway) and then stored in TRIzol® Reagent (Ambion™, Life Technology, Carlsbad, USA) at −80 °C. Total RNA, including miRNA, was isolated using TRIzol ® Reagent according to manufacturer’s instructions. Quality and concentration of the isolated RNA were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA); all samples showed an A260/280 ratio of >1.80.
miRNA profiling and identification
Agilent’s miRNA microarray system was used to measure miRNA. Total RNAs were prepared and subjected to a quality check using the bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). An 8×12 K miRNA microarray (Agilent Technologies, Santa Clara, CA, USA) was used to visualize the expression profiles of miRNAs. All procedures were carried out according to the manufacturer’s instructions. The expressed miRNA data were normalized using the quantile normalization method. The threshold used to screen up- or down-regulated miRNAs was a fold change (FC) of ≥1.5 and P value <0.05.
Quantitative assessment of miRNA
A 600 ng of total RNA was reverse transcribed by Mir-X miRNA First-Strand Synthesis Kit (TAKARA). Quantitative polymerase chain reaction (qPCR) was performed on a StepOnePlus RealTime PCR System (Applied Biosystems) in a 20 µL reaction that contained 1 µL of cDNA and Power SYBR® Green PCR Master Mix (Applied Biosystems). The expression level of U6 was used as control. The relative expression was calculated with the comparative Ct method and was expressed as the FC compared to the control. Three replicates were made for each sample. Besides primers of U6 were provided in the TAKARA Synthesis Kit, other miRNA primers were using the following sequence: hsa-miR-194-3p 5’-ATTATTCCAGTGGGGCTGCT-3’, hsa-miR-495-3p 5’-GCCGTCAAACAAACATGGTG-3’ and hsa-miR-223-5p 5’-GCCTGCCGTGTATTTGACAA-3’.
Statistical analysis
All experiments were conducted independently with at least three replicates. Data were presented as median (range) for age, as number for categorical variables and mean ± standard deviation (SD) for continuous variables. Data were analyzed using SPSS (version 13.0, SPSS Inc., Chicago, USA). Comparisons between two groups were estimated by two-tailed Student’s test or Mann-Whitney test, except that comparisons of lung function test in time-serious study were estimated by paired t-test. Spearman correlation analyses were taken to evaluate the relationship between miRNAs and lung function results. P<0.05 was considered statistically significant.
Results
miRNAs profiles and validation
Differentially expressed miRNAs were identified from the discovery study comparing FEV1 decline and control group and the result was submitted in GEO (GSE102915). In total, 21 miRNAs were differentially expressed in FEV1 decline individuals with 18 upregulation and 3 downregulation (Figure 2A). The location and the number of predicted genes were also shown.
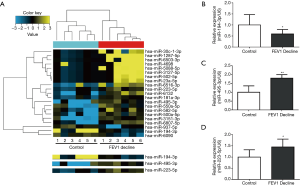
Expression of miR-495-3p, miR-223-5p and miR-194-3p were further validated by qRT-PCR. miR-495-3p was chosen because it significantly increased in FEV1 decline group, while miR-194-3p was chosen cause its dramatic decreasing in expression. As miR-223 has been reported to be an important regulator in COPD (14,15), miR-223-5p was also chosen for validation. The relative expression of miR-495-3p, miR-223-5p and miR-194-3p showed significant difference between control group and FEV1 decline group (Figure 2B,C,D).
Pulmonary ventilation function before and after exposure
Pulmonary ventilation function results at lag0 and lag1 day were analyzed (Figure 3A,B). At lag1 day, predicted FEV1% (88.21%±10.57%, P<0.007) and predicted FVC% (93.83%±10.82%, P<0.001) decreased significantly in “after exposure”, compared with predicted FEV1% (91.10%±10.80%) and predicted FVC% (97.45%±10.41%) in “before exposure”. Similar at lag0 day, predicted FEV1% (92.40%±9.95% before vs. 89.93%±10.56% after, P<0.009) and predicted FVC% (99.38%±10.13% before vs. 95.75%±10.07% after, P<0.002) decreased significantly. However, FEV1/FVC shown no statistical difference neither at lag1 nor lag0.
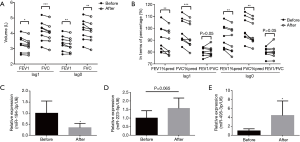
Expression of miR-495-3p, miR-223-5p and miR-194-3p before and after exposure
In the time-series study, expression of miR-495-3p, miR-223-5p and miR-194-3p were measured by qRT-PCR (Figure 3C,D,E). Compared with “before exposure”, expression of miR-495-3p (P=0.032) and miR-223-5p (P=0.048) increased statistically in blood samples of “after exposure”. Expression of miR-194-3p (P=0.007) showed a significant decrease after exposure.
Correlation between differentially expressed miRNAs and lung function
The relationship between miRNAs and lung function were measured respectively in time-serious study at lag0 and lag1. miR-194-3p showed statistically positive correlation with predicted FEV1% (P=0.037) and predicted FVC% (P=0.023) both at lag1 and lag0. However, neither miR-495-3p nor miR-223-5p showed a statistically relationship with lung ventilation function results.
Discussion
In this pilot study, firstly, we measured miRNA expression in blood from COPD participants and control participants. We identified a set of 21 miRNAs being differentially expressed. And we specifically focused on three of the 21 miRNAs of the discovery cohort including miR-194-30, miR-495-3p and miR-223-5p. miR-194-3p and miR-495-3p were the most decreased and increased miRNA respectively. There were very limited evidences of their function, and they might be a novel biomarker in the context of lung dysfunction. Expression miR-194-3p was reported to be altered in chronic hepatitis B infection (16) and necrotizing enterocolitis and spontaneous intestinal perforation of infants (17). However, the function of miR-194-3p is still not known. An in vitro study demonstrated decreased miR-194 was related to increased secretion of extracellular matrix proteins resulting in hepatic fibrosis (18). On the other hand, miR-495-3p has a potential to be a tumor suppressor. Increasing expression of plasma miR-495-3p was associated with effective radio-sensitivity in patients with non-small cell lung cancer (19), and suppression of miR-495-3p induced overexpression of tumor cell growth and proliferation related oncogenic epigenetic modifiers in gastric carcinogenesis (20).
Besides, miR-223-5p is believed to be an essential regulator in the pathogenesis of COPD. Compared with smokers without airway obstruction, expression of miR-223 increased in the lung tissue of smokers with COPD (14). And miR-223 plays an essential role in promoting granulocytic differentiation while also being associated with the suppression of erythrocytic differentiation. Upregulated miR-223 increased myeloid cells recruitment in lung by directly targeting the chemoattractants in myeloid cells, and consequently, introduced neutrophil-driven inflammation (21).
Next, in order to figure out the influence of PM2.5 on these three miRNAs, we validated miR-194-3p, miR-495-3p and miR-223-5p in a time-series study. The results showed that blood expression of miR-194-3p decreased whereas miR-223-5p increased after exposure of PM2.5. Before, miR-223 has been reported to be associated with environmental exposure. Maternal tobacco smoke exposure during pregnancy correlated with increased expression of maternal blood miR-223 and lower cord blood Treg cell numbers, which increased the allergy risk of infants (22). Furthermore, although miR-194-3p showed marginally significance in COPD, we found decreased miR-194-3p was highly associated with lung ventilation function decline in the time-series study. Reduced FEV1 and FVC were high risk factors of pulmonary diseases such as COPD (4). The mechanism of pulmonary ventilation function is a huge and complicate biological process including inflammation, oxidative stress, cell proliferation, apoptosis and cell repair (6,23-25). In vitro, we found PM2.5 significantly aggravated apoptosis in cigarette-inflamed human bronchial epitheliual cells, and miR-194-3p was dramatically downregulated in PM2.5 plus cigarette smoke solution treated cells. Importantly, overexpression of miR-194-3p could suppressed the apoptotic response (26).
As this was a pilot study, there were some inevitable limitations. Firstly, miRNAs are reported to be sensitive indicators of the effects of acute and chronic environmental exposure. However, there are little overlap among in resent studies because of the diversity of design (9). Though we designed a two-step study to narrow down the target miRNAs, there existed some influence factors not taken into account. Taking as an example, expression of miR-223 is higher in COPD smokers than smokers without obstruction (14), and smoking exposure might be associated with expression of miR-223 (22). It indicated that state of airway disease should be another influence factor. Secondly, the study population was limited. Thus, formal and large-scale study should be designed to observe the role of different miRNAs like miR-194-3p and miR-223-5p in people with different smoking status and disease state. Additionally, it is still a huge challenge to give information of the PM2.5 levels that these individuals were exposed. In order to reduce the influence of different individual exposure level as possible, we tried to consider include criteria carefully in the time-series study. All chosen participants in discovery study were Beijing residents with outdoor activities per day. The lag0–2 air quality index (AQI) were <150 in control group and <100 in lung function decline group. These AQI levels were recommended with small influence on healthy person and pulmonary disease respectively by Beijing Municipal Environmental Protection Bureau (27). And volunteers in time-series study basically shared the same daily life track.
Conclusions
Blood miR-495-3p, miR-223-5p and miR-194-3p was screened to be COPD associated miRNAs. After confirmation in a time-series study, blood miR-194-3p decreased and miR-223-5p increased significantly after exposure of PM2.5. And miR-194-3p was highly positive correlated with lung ventilation function during exposure of PM2.5. These results indicated that miR-194-3p might be a protective biomarker in PM2.5-triggered pulmonary dysfunction such as COPD. The present result might provide new clues for further studies to understand the mechanism of these miRNAs in both environmental exposure and COPD. However, due to the limited study population, further investigations are necessary.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Bureau BMEP. Beijing Daily Air Quality Report. Available online: http://www.bjepb.gov.cn/bjhrb/zrqskqzl/index.html?from=groupmessage&isappinstalled=0
- Bureau BMEP. Beijing Environmental Statement 2015. Available online: http://hbdc.mep.gov.cn/hbdt/bjdt/201612/t20161219_371176.shtml
- Atmospheric aerosols: composition, transformation, climate and health effects. Angew Chem Int Ed Engl 2005;44:7520-40. [Crossref] [PubMed]
- GOLD. Global Strategy for the Diagnosis, Management, and Prevention of COPD: Full Repo (2017). Available online: http://goldcopd.org
- MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Role of apoptosis in the pathogenesis of COPD and pulmonary emphysema. Respir Res 2006;7:53. [Crossref] [PubMed]
- miR-146a-5p plays an essential role in the aberrant epithelial-fibroblast cross-talk in COPD. Eur Respir J 2017.49. [PubMed]
- Effects of PM2.5 exposure on the Notch signaling pathway and immune imbalance in chronic obstructive pulmonary disease. Environ Pollut 2017;226:163-73. [Crossref] [PubMed]
- MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect 2015;123:399-411. [Crossref] [PubMed]
- Elevated expression of miR-146, miR-139 and miR-340 involved in regulating Th1/Th2 balance with acute exposure of fine particulate matter in mice. Int Immunopharmacol 2018;54:68-77. [Crossref] [PubMed]
- Short-term effects of fine particulate matter exposure on pulmonary function in healthy young adults: a time series study (Chinese). Zhonghua Yi Xue Za Zhi 2017;97:1430-5. [PubMed]
- Association between changes in air pollution levels during the Beijing Olympics and biomarkers of inflammation and thrombosis in healthy young adults. Jama 2012;307:2068-78. [Crossref] [PubMed]
- Cardiorespiratory biomarker responses in healthy young adults to drastic air quality changes surrounding the 2008 Beijing Olympics. Res Rep Health Eff Inst 2013;(174).5-174. [PubMed]
- Gene expression networks in COPD: microRNA and mRNA regulation. Thorax 2012;67:122-31. [Crossref] [PubMed]
- Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res 2014;15:149. [Crossref] [PubMed]
- The expression of miR-125b-5p is increased in the serum of patients with chronic hepatitis B infection and inhibits the detection of hepatitis B virus surface antigen. J Viral Hepat 2016;23:330-9. [Crossref] [PubMed]
- Comparative MiRNA Expressional Profiles and Molecular Networks in Human Small Bowel Tissues of Necrotizing Enterocolitis and Spontaneous Intestinal Perforation. PLoS One 2015;10:e0135737. [Crossref] [PubMed]
- Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol 2010;298:G101-6. [Crossref] [PubMed]
- Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol 2016;37:11927-36. [Crossref] [PubMed]
- MicroRNA-495-3p functions as a tumor suppressor by regulating multiple epigenetic modifiers in gastric carcinogenesis. J Pathol 2018;244:107-19. [Crossref] [PubMed]
- MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest 2013;123:4836-48. [Crossref] [PubMed]
- Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol 2014;133:543-50. [Crossref] [PubMed]
- Epithelial repair mechanisms in the lung. Am J Physiol Lung Cell Mol Physiol 2010;298:L715-31. [Crossref] [PubMed]
- Increased phosphorylated p38 mitogen-activated protein kinase in COPD lungs. Eur Respir J 2013;42:28-41. [Crossref] [PubMed]
- Proinflammatory cytokines induce bronchial hyperplasia and squamous metaplasia in smokers: implications for chronic obstructive pulmonary disease therapy. Am J Respir Cell Mol Biol 2012;47:67-79. [Crossref] [PubMed]
- PM2.5 downregulates miR-194-3p and accelerates apoptosis in cigarette-inflamed bronchial epithelium by targeting death-associated protein kinase 1. Int J Chron Obstruct Pulmon Dis 2018;13:2339-49. [Crossref] [PubMed]
- Bureau BMEP. Air Quality Index (AQI). Available online: http://www.bjmemc.com.cn/gzdt_getOneInfo.action?infoID=1662


