
The landscape of early carcinogenesis revealed through the lens of integrative genomics, epigenomics, and transcriptomics
Molecular signatures in early carcinogenesis: an unmet need
The hallmarks of established cancers have been meticulously catalogued over decades, almost entirely through the study of advanced disease (1). By contrast, the molecular features of early pre-neoplastic lesions including the occult alterations that presage progression to invasive and metastatic cancer, remain elusive (2). A better understanding of the biology of nascent cancers and the complex genetic and epigenetic evolutionary processes that drives them towards lethal disease, represents an important step towards more effective early intervention strategies.
In the absence of reliable molecular profiling data, histo-morphologic architecture has been the default method for classifying pre-invasive lesions. The resulting grading systems describe a step wise progression with each sample offering a single cross sectional picture in time (3,4). However, the evolution of early disease is not unidirectional; spontaneous regression and involution of dysplastic lesions is not only possible but may indeed occur more often than appreciated (5,6). Moreover, tissue and cellular morphology alone are only weak predictors of the capacity of pre-invasive lesions to progress to invasive disease (7). In recent years, with the advent and widespread use of molecular profiling, it has become apparent that neoplastic tissues harbor complex genetic and epigenetic variegation. Whether or not such alterations have prognostic or predictive value that can be leveraged to guide the management of pre-invasive lesions remains an open question.
Analysis of pre-invasive lung cancer reveals the occult signature of future progression
In the March 2019 edition of Nature Medicine, Teixeira et al. report an unprecedented molecular analysis of pre-invasive lung cancer in a prospectively tracked cohort of patients with bronchial carcinoma in situ (CIS), a precursor to invasive squamous cell carcinoma (SCC) (8). This multi-national group of investigators performed serial bronchoscopies and biopsies, tracking the evolution of airway CIS lesions in a longitudinally monitored cohort. Although microscopically indistinguishable from one another, approximately half of the CIS lesions in this cohort remained static or spontaneously regressed while the other half progressed to invasive SCC, consistent with previous reports describing the natural history of bronchial CIS (9,10). At the time of regression or progression, they profiled the genomic, transcriptomic, and epigenomic landscape of the index CIS lesion and identified progression-specific methylation changes and chromosomal instability (CIN) signatures within the heterogenous molecular background of these lesions. In addition, mutations and copy number changes characteristic of cancer were charted, offering a window into the evolutionary dynamics of early carcinogenesis. In total, 129 index CIS biopsies were obtained from 85 patients and underwent pathologic and molecular characterization. Remarkably, predictive modeling reliably identified the lesions destined to progress with a high degree of specificity (Figure 1).
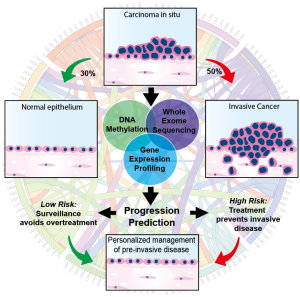
Genomic profiling of progressive samples showed frequent alterations in putative driver genes including TP53, CDKN21, SOX2, AKT. Less frequent alterations were seen in FAT1, KMT2D, KEAP1, EGFR and NOTCH1. By contrast, regressive samples had a lower mutational burden and fewer copy number alterations. Transcriptomic and epigenomic profiles revealed 1,135 genes associated with CIS fate. Of these genes TPM3, PTPRB, SLC34A2, KEAP1, NKX2-1, SMAD4, and SMARCA4 were identified as potential drivers of invasive progression. Homeobox family genes (HOXC8, HOXC9, HOXC10, HOXD10, HOXA11AS), which are coordinately expressed during development, epigenetically silenced, and often reactivated in early carcinogenesis, were highly enriched in the differential expression and methylation analyses. Similarly, the NKX2-1 gene was frequently hypermethylated and transcriptionally silenced in progressive samples. Invasive progression was strongly associated with a CIN signature (i.e., gain or loss of whole/part of a chromosome). CIN70 signature genes ACTL6A, ELAVL1, MAD2L1, NEK2, OIP5 were noted to be upregulated in progressive samples but not in regressive samples.
Implications for biomarker development targeting early carcinogenesis
Efforts to probe the primitive clonal events in early carcinogenesis promise to expand avenues towards detecting and eradicating cancers at their vulnerable nascent stages. Previous studies of precursor biology in a variety of cancers have similarly identified genetic and epigenetic alterations as early events in tumor development (11-15). Profiling studies in breast, prostate, bladder, colorectal, and other cancers have afforded insights into early tumor evolution and spurred the development of predictive and prognostic biomarkers, some of which have gone forward to validation studies and clinically utilization (16-20). Additional biomarkers are urgently needed to improve clinical risk stratification at earlier stages with the goal of tailoring therapies to an individual’s tumor biology. In lung cancer, effective local therapies such as surgery and stereotactic radiotherapy are available, however they carry risks and appropriate patient selection using clinical criteria can be challenging (21-23). Teixeira and colleagues’ pioneering contribution in this arena and has potential to improve early detection, reduce overtreatment, select high risk patients for curative local therapy, and cultivate new interventions and prevention strategies. The underlying premise of this work has broad applicability in oncology and similar investigations in other pre-invasive thoracic and extra-thoracic malignancies are needed. Furthermore, biomarker informed clinical trials powered to validate the findings of such studies are worthy of attention and investment from clinical cooperative groups.
The study of early carcinogenesis: looking forward
Open questions remain in the quest to better understand the biology and predict the progression of pre-invasive lesions. Current molecular profiling methods are biased toward tumor-intrinsic features and underestimate the important influence of a lesion’s microenvironment (24). Similarly, sampling limitations complicate the assessment of rare variants, regional heterogeneity, and field cancerization effects, all of which undoubtedly have implications for prognosis and treatment (25). Moreover, while promising studies like the work of Teixeira et al will allow us to better predict the fate of sampled lesions, they do not address the optimal strategy to modify the natural history of an individual’s pre-invasive disease. Both prognostic and better predictive biomarkers will be needed to guide management of lesions with aggressive biology, e.g., by resection, radiation, focal ablation, or systemic treatment including targeted approaches, epigenetic therapy, and immunomodulatory strategies. As improved imaging, molecular profiling, effective treatments, and the pipeline of emerging biomarkers converge to focus on ever earlier stages of disease, there is good reason for patients and clinicians to be optimistic about the future of thoracic oncology.
Acknowledgments
Funding: OY Mian is supported by DoD, CDMRP, W81XWH-18-1-0177; NCI NIH HHS L30 CA220908; Case Comprehensive Cancer Center, P30 CA043703; VeloSano Foundation. ME Abazeed is supported by NIH KL2 TR0002547, NIH R37 CA222294, the American Lung Association and VeloSano Foundation.
Footnote
Conflicts of Interest: ME Abazeed reports the following COI: Siemens Healthcare: Grant Support; Bayer AG: Grant Support, Travel Support, Honorarium. The other authors have no conflicts of interest to declare.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Thakrar RM, Pennycuick A, Borg E, et al. Preinvasive disease of the airway. Cancer Treat Rev 2017;58:77-90. [Crossref] [PubMed]
- Rami-Porta R, Bolejack V, Goldstraw P. The new tumor, node, and metastasis staging system. Semin Respir Crit Care Med 2011;32:44-51. [Crossref] [PubMed]
- Zugazagoitia J, Enguita AB, Nuñez JA, et al. The new IASLC/ATS/ERS lung adenocarcinoma classification from a clinical perspective: current concepts and future prospects. J Thorac Dis 2014;6:S526-36. [PubMed]
- Salman T. Spontaneous tumor regression. J Oncol Sci 2016;2:1-4.
- Challis GB, Stam HJ. The Spontaneous Regression of Cancer: A review of cases from 1900 to 1987. Acta Oncol 1990;29:545-50. [Crossref] [PubMed]
- Kadota K, Nitadori J, Rekhtman N, et al. Reevaluation and reclassification of resected lung carcinomas originally diagnosed as squamous cell carcinoma using immunohistochemical analysis. Am J Surg Pathol 2015;39:1170-80. [Crossref] [PubMed]
- Teixeira VH, Pipinikas CP, Pennycuick A, et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med 2019;25:517-25. [Crossref] [PubMed]
- Merrick DT, Gao D, Miller YE, et al. Persistence of Bronchial Dysplasia Is Associated with Development of Invasive Squamous Cell Carcinoma. Cancer Prev Res (Phila) 2016;9:96. [Crossref] [PubMed]
- Breuer RH, Pasic A, Smit EF, et al. The Natural Course of Preneoplastic Lesions in Bronchial Epithelium. Clin Cancer Res 2005;11:537-43. [PubMed]
- Mian OY, Wang SZ, Zhu SZ, et al. Methyl-binding domain protein 2-dependent proliferation and survival of breast cancer cells. Mol Cancer Res 2011;9:1152-62. [Crossref] [PubMed]
- Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nat Rev Cancer 2015;15:473-83. [Crossref] [PubMed]
- Hitchins MP. Constitutional epimutation as a mechanism for cancer causality and heritability? Nat Rev Cancer 2015;15:625-34. [Crossref] [PubMed]
- Mian OY, Khattab MH, Hedayati M, et al. GSTP1 Loss results in accumulation of oxidative DNA base damage and promotes prostate cancer cell survival following exposure to protracted oxidative stress. Prostate 2016;76:199-206. [Crossref] [PubMed]
- Ryan BM, Faupel-Badger JM. The hallmarks of premalignant conditions: a molecular basis for cancer prevention. Semin Oncol 2016;43:22-35. [Crossref] [PubMed]
- Clark SE, Warwick J, Carpenter R, et al. Molecular subtyping of DCIS: heterogeneity of breast cancer reflected in pre-invasive disease. Br J Cancer 2011;104:120-7. [Crossref] [PubMed]
- Mian OY, Tendulkar RD, Abazeed ME. The evolving role of molecular profiling in prostate cancer: basal and luminal subtyping transcends tissue of origin. Transl Cancer Res 2017;6:S1441-5. [Crossref] [PubMed]
- Koshkin VS, Garcia JA, Reynolds J, et al. Transcriptomic and Protein Analysis of Small-cell Bladder Cancer (SCBC) Identifies Prognostic Biomarkers and DLL3 as a Relevant Therapeutic Target. Clin Cancer Res 2019;25:210-21. [Crossref] [PubMed]
- Batista da Costa J, Gibb EA, Bivalacqua TJ, et al. Molecular characterization of neuroendocrine-like bladder cancer. Clin Cancer Res 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Tran NH, Cavalcante LL, Lubner SJ, et al. Precision medicine in colorectal cancer: the molecular profile alters treatment strategies. Ther Adv Med Oncol 2015;7:252-62. [Crossref] [PubMed]
- Woody NM, Stephans KL, Andrews M, et al. A Histologic Basis for the Efficacy of SBRT to the lung. J Thorac Oncol 2017;12:510-9. [Crossref] [PubMed]
- Stephans KL, Woody NM, Reddy CA, et al. Tumor Control and Toxicity for Common Stereotactic Body Radiation Therapy Dose-Fractionation Regimens in Stage I Non-Small Cell Lung Cancer. Int J Radiat Oncol Biol Phys 2018;100:462-9. [Crossref] [PubMed]
- Manyam BV, Videtic GMM, Verdecchia K, et al. Effect of Tumor Location and Dosimetric Predictors for Chest Wall Toxicity in Single-Fraction Stereotactic Body Radiation Therapy for Stage I Non-Small Cell Lung Cancer. Pract Radiat Oncol 2019;9:e187-95. [Crossref] [PubMed]
- Brock A, Krause S, Ingber DE. Control of cancer formation by intrinsic genetic noise and microenvironmental cues. Nat Rev Cancer 2015;15:499-509. [Crossref] [PubMed]
- Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat Rev Cancer 2018;18:19-32. [Crossref] [PubMed]


