
Comprehensive analysis of EGFR T790M detection by ddPCR and ARMS-PCR and the effect of mutant abundance on the efficacy of osimertinib in NSCLC patients
Introduction
In recent years, epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) have been largely successful for the clinical treatment of patients with non-small cell lung cancer (NSCLC) harboring activating EGFR mutations. Most of the mutations associated with sensitivity to anilinoquinazoline EGFR inhibitors occur as either multi-nucleotide in-frame deletions in exon 19 (19del), eliminating four amino acids, Leu-Arg-Glu-Ala, or as a single nucleotide variation at nucleotide 2573 (T to G) in exon 21, resulting in substitution of arginine for leucine at position 858 (L858R) (1,2). However, a majority of patients eventually acquire resistance to the drug, with a median time to progression of 10 to 14 months (3,4). Cases where patients with mutations, such as 19del, L858R, G719X, or L861Q (known to be associated with TKI sensitivity), develop systemic progression of disease while on treatment with EGFR-TKIs is defined as acquired resistance to EGFR-TKIs (5).
The T790M mutation leads to threonine-to-methionine amino acid change at position 790 of the EGFR tyrosine kinase domain, causing steric hindrance that may interfere with the binding of TKIs (6). It occurs in 50% to 60% of patients undergoing treatment with EGFR-TKIs and is the most frequent alteration leading to acquired resistance (7-9). In addition, de novo T790M mutation is also an important mechanism of primary resistance to EGFR-TKIs (10). The highly sensitive methods of droplet digital polymerase chain reaction (ddPCR) and amplification refractory mutation system (ARMS)-PCR are routinely applied in clinical detection of T790M mutation (11,12). In this study, we compared the detection rates of these two methods and analyzed the associations of T790M status with clinicopathological parameters and progression-free survival (PFS) in patients with NSCLC, providing detailed evidence to better inform clinical decision-making and improve outcomes.
Methods
Patients
From August 2017 to February 2019, 263 cases that consulted for T790M mutation test by ddPCR in the department of molecular diagnostics of Sun Yat-sen University Cancer Center were retrospectively collected. All patients were diagnosed with NSCLC by pathological examination and the last follow-up was done on 26th February 2019. Objective tumor responses were evaluated every 6–8 weeks in accordance with the Response Evaluation Criteria in Solid Tumors guidelines (version 1.1) (13). Patients with sensitive EGFR mutation had received erlotinib, gefitinib or icotinib orally at a recommended dose, and some patients had received osimertinib treatment. The current study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center, and all patients provided signed informed consent.
DNA extraction
Genomic DNA (gDNA) was extracted from formalin fixed paraffin-embedded (FFPE) tumor tissue and cell pellet centrifuged from hydrothorax using a QIAGEN DNA FFPE Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions and quantified with a Nano-Drop2000 (NanoDrop Technologies, Wilmington, DE, USA). From 10 mL of whole blood, 5 mL plasma was collected and used to isolate and purify circulating tumor DNA (ctDNA) using a QIAamp Circulating Nucleic Acid Kit (Qiagen), following the manufacturer’s instructions.
ARMS-PCR and ddPCR
ARMS assay (AmoyDx, Xiamen, China) was conducted using ABI 7500 (Applied Biosystems, Foster City, CA, USA), while ddPCR assay (YUANQI BIO, Shanghai, China) was performed by QX200 Droplet Digital PCR (ddPCR™) (BIO-RAD, Hercules, CA, USA) system. The result was interpreted as positive when the mutant copy number ≥3 in ddPCR, and the T790M abundance was calculated as 100%× (mutant copy number/total copy number).
Statistical analysis
PFS1 was defined as the time from the start of the first-generation EGFR-TKI treatment to the first documentation of progressive disease (PD) or the last follow-up, and PFS2 was defined as the time from the beginning of osimertinib treatment to the second PD or the last follow-up. All time-to-event outcomes were estimated using the Kaplan-Meier method and compared across groups using the log-rank test. The associations between T790M and clinical characteristics were analyzed using the Chi-squared test. Differences between groups were assessed by Student’s t-test or one-way analysis of variance. The cut-off value of T790M mutant abundance was decided using the receiver operating characteristic curve. Two-tailed P values <0.05 were considered statistically significant. SPSS 23.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 6 (La Jolla, CA, USA) software were used for statistical analyses and graphical representations.
Results
Clinicopathological characteristics of patients
As shown in Table 1, samples of 115 males and 148 females were included in our study, and most of them were diagnosed as adenocarcinoma in TNM stage IV. Sample types included tissue, hydrothorax, and peripheral blood (PB). The average age of the patients was 59.5 (ranging from 26 to 87). Eighty-eight patients had 19del, 87 patients had L858R, 53 patients had mutations of other types, and 35 patients were classified as wild type (WT). Among all cases, 203 patients had received first-generation TKIs as first-line treatment, and 68 patients had received osimertinib after the first PD.
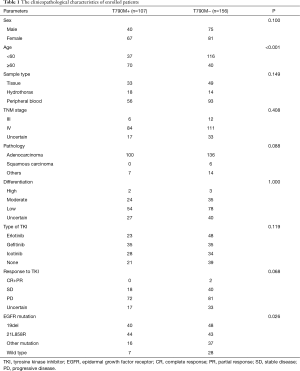
Full table
Comparison of detection efficacy of ddPCR and ARMS PCR
In our study, the total positive rate of T790M in 263 samples tested by ddPCR was 40.7% (107/263), the mean mutant abundance and copy number were 8.1% (0.06% to 76.3%) and 241 (3 to 2,916), respectively. Along with ddPCR, 53 samples were also subjected to ARMS-PCR at the same time, of which there were 18 samples judged negative by both ddPCR and ARMS-PCR. However, only 21 samples were recognized as positive by ARMS-PCR, as opposed to 35 positive samples detected by ddPCR (Table 2). The average T790M mutant abundance and copy number in the ddPCR+ARMS+ group (19.1%, 636.9) were both significantly higher than that in the ddPCR+ARMS− group (0.36%, 12.1) (Figure 1A,B). Noticeably, the sensitivity and the concordant rate (true negative and true positive) of ARMS-PCR were only 27.8% and 63.9% of ddPCR in samples with mutant abundance of <1%, and they gradually added up to 60.0% and 73.6%, respectively, when the mutant abundance range was enlarged to <100% (Figure 1C,D).
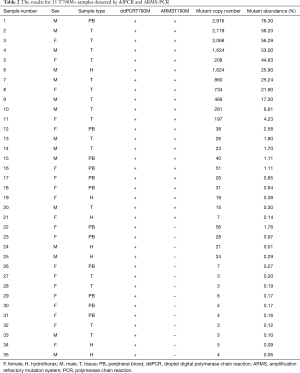
Full table
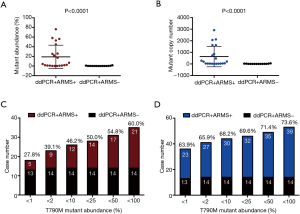
Together, it demonstrated that both ddPCR and ARMS-PCR exhibited a high specificity in detection of negative samples. However, ddPCR performed better than ARMS-PCR in identifying positive samples and was especially sensitive in detecting less abundant T790M mutation.
Influence of sample type on detection rate of T790M by ddPCR
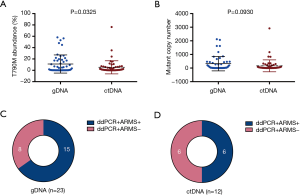
According to our analysis, the T790M positive rates were 40.2% (33/82), 56.3% (18/32), and 37.6% (56/149) in tissue, hydrothorax, and PB sample groups, respectively. In terms of DNA source, the T790M detection rate in ctDNA from plasma of PB was similar to that in gDNA from tissue and cells of hydrothorax (37.6% vs. 44.7%, P=0.242). The average mutant abundance in T790M+ gDNA samples was statistically higher than that in ctDNA samples (11.1% vs. 5.3%, P=0.0325). Nevertheless, the average mutant copy number in T790M+ gDNA samples was numerically but not statistically higher than that in ctDNA samples (323.8 vs. 165.3, P=0.0930) (Figure 2A,B). The ddPCR+ARMS+/ddPCR+ARMS− ratio in gDNA samples (15/8) was also numerically, but not statistically higher than that in ctDNA samples (6/6, P=0.383) (Figure 2C,D).
Interestingly, among three patients (A, B and C) who had submitted both tissue and PB samples to be tested by ddPCR, patient A got consistent negative results, patient B got consistent positive results, and it is worth noticing that the mutant abundance in tissue sample (58.2%) was much higher than that in PB sample (10.8%). Patient C tested positive on the basis of the tissue sample (14.9%), but negative on the basis of the PB sample (0%). In addition, another two patients (D and E) had submitted both hydrothorax and PB samples to be measured by ddPCR; patient D was judged as T790M− in both samples and patient E was judged as T790M+ in both samples, but with different mutant abundances—6.9% in hydrothorax and 1.1% in PB (Table 3).
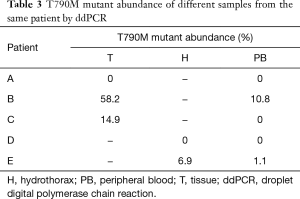
Full table
Accompanying sensitive mutation composition in pre- and post-TKI subset
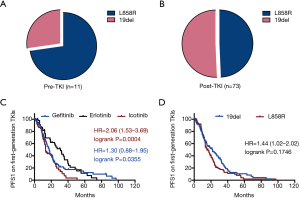
T790M mutation is one of the most important causes of primary TKI resistance (10). In the current study, T790M+ was detected in 11 patients before they received any TKI therapies. These patients comprised six males and five females, and they submitted eight tissue samples and three hydrothorax samples. We observed that primary T790M mutation occurred with L858R in eight samples and 19del in three samples in the pre-TKI subset, while acquired T790M coexisted with 19del in 37 samples and L858R in 36 samples in the post-TKI subset (Figure 3A,B, Table S1).

Full table
PFS1 of patients with sensitizing EGFR mutation receiving first-generation TKIs
PFS1 was assessed in 203 patients receiving the first generation of TKIs based on TKI type and activating mutation type. According to the Kaplan-Meier survival analysis, the PFS1 of erlotinib-group was markedly longer than that of the gefitinib-group (31.0 vs. 17.5 months, P=0.0355) and that of icotinib-group (31.0 vs. 13.2 months, P=0.0004) (Figure 3C), while the PFS1 was similar in patients with 19del versus L858R mutations (21.9 vs. 15.2 months, P=0.1746) (Figure 3D).
T790M mutation and its mutant abundance predicts osimertinib therapeutic effect
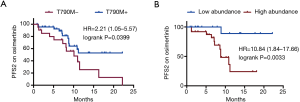
Among the assessable patients, 47 T790M+ patients and 21 T790M− patients had received osimertinib treatment after PD. Among them, 43 patients were evaluated as stable disease (SD) and 25 patients were evaluated as PD until the last follow-up. The data showed that the PFS2 in T790M+ patients was significantly longer than that in the T790M− patients on osimertinib (not achieved vs. 10.1 months, P=0.0399) (Figure 4A). Moreover, patients with T790M abundance lower than 1.065% had a more prolonged PFS2 than those with higher T790M abundance (not achieved vs. 8.8 months, P=0.0033, Figure 4B).
Discussion
Newly acquired resistance in NSCLC patients with activating EGFR mutation after first-generation TKI therapy is a serious problem leading to poor outcome of the disease. The secondary T790M mutation in the catalytic cleft of the EGFR tyrosine kinase domain introduces a bulkier amino acid side chain in the ATP-kinase-binding pocket, which alters the activity of erlotinib and gefitinib by increasing the ATP affinity at the binding pocket, thereby minimizing the efficacies of the EGFR-TKIs (6,14). According to our analysis, the total detection rate of T790M in all types of samples was 40.7%, which was similar to the detection rate of about 50% reported in re-biopsy tumor tissue from NSCLC patients with acquired resistance to EGFR-TKI therapy (9).
Our data showed that ddPCR was extremely sensitive in detecting T790M with low mutant abundance, and this advantage was supported by several other studies. Wang et al. demonstrated that the T790M detection rates were 46.7% (35/75) and 25.3% (19/75) in 75 patient plasma samples by ddPCR and ARMS-PCR, respectively (12). Feng et al. showed that only 3 out of 17 T790M+ samples were identified as positive by ARMS-PCR, and none of the T790M+ samples with mutant allele frequency <0.5% was correctly confirmed by ARMS-PCR (15). Additionally, Zhang et al. reported that the results for 9 out of 10 clinical samples were consistent by ARMS-PCR and ddPCR; however, one sample indicated to be EGFR WT by ARMS-PCR was confirmed as T790M+ with 7 mutant copies in the background of 6,000 WT copies (abundance 0.12%) by ddPCR (11). In the present study, 18 T790M− samples were all classified as negative by both methods; however, 14 T790M+ samples with abundance <2.0% were inconsistently recognized as EGFR WT by ARMS-PCR, illustrating that ddPCR has a parallel specificity with and a much higher sensitivity than ARMS-PCR. Therefore, for potential beneficiaries of osimertinib with low level of T790M mutation, which is difficult to be recognized by ARMS-PCR, ddPCR would be a more advisable choice to avoid false negative results.
The choice of sample type is always an important concern faced by clinicians. Currently, tissue sample remained the optimized choice when available, considering its higher DNA yield and mutation content. However, re-biopsy cannot be routinely obtained in the clinical practice. Previous studies have shown the feasibility of investigating EGFR mutation status in ctDNA using different technologies; the detection rate ranged from 16.7% to 34.3%, and concordance rate ranged from 66.3% to 92.9% (16-18). In our study, the detection rate in ctDNA using ddPCR was 37.6%, which was comparable with that in gDNA (44.7%). Besides, PB sample is relatively easy to get during the whole treatment process and ctDNA could reflect the dynamic change of disease progression. As more molecularly tailored treatment options become available for NSCLC patients, ctDNA in PB sample should become a generally accepted source of DNA, providing a potential alternative to tumor-derived samples for EGFR mutation analysis.
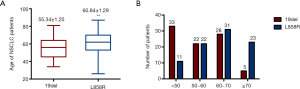
T790M was reported to always occur concurrently with sensitizing EGFR mutations, and we found 92.5% (99/107) T790M+ patients harbored sensitizing EGFR mutations. It has been established that acquired T790M was more prevalent to coexist with 19del, while primary T790M mutation was more likely to coexist with L858R. Some rare second-site mutations implicated in acquired resistance other than T790M, such as D761Y, L747S, and T854A, were reported to always coexist with L858R (7,19-21). Wang et al. reported that 31 patients with 19del and 14 patients with L858R harbored acquired T790M (12). Li et al. reported that 44 patients with 19del and 25 patients with L858R harbored acquired T790M, while 10 patients with 19del and 30 patients with L858R harbored primary T790M (22). A meta-analysis showed that T790M was more frequent in 19del than in L858R among patients with acquired resistance to TKIs (53% vs. 36%; OR 1.87; P<0.001) (23). Another meta-analysis suggested that primary T790M is less frequent in patients harboring 19del compared with those carrying L858R (14% vs. 22%; OR 0.59; P<0.001) (24). In this study, we found that 37 patients with 19del and 36 patients with L858R carried acquired T790M, 3 patients with 19del and 8 patients with L858R carried primary T790M (Figure 3A,B, Table S1), which is roughly consistent with previous data. However, the ratio of 19del to L858R in acquired T790M group was not that high in this study compared with the results reported in the above-mentioned studies. Zhuo et al. reported that 21.4% patients were aged 65 years or above in the 19del group, whereas this proportion was markedly higher in the L858R group (38.4%, P=0.015) (25), suggesting that the mutation type might be associated with age. In light of this report, we analyzed our data and found that the average age of patients with L858R was significantly higher than that of patients with 19del (60.8 vs. 55.3, P=0.0029, Figure S1A). Moreover, there were considerably more patients older than 70 years old and less patients younger than 50 years old in the L858R group, compared with the 19del group (Figure S1B), suggesting that the difference in age distribution might be one of the reasons why acquired T790M tended to frequently coexist with L858R in this study.
In addition, some researchers have speculated that 19del might play a distinct biological function considering its higher coincidence with acquired T790M. Preclinical data have demonstrated that the EGFR T790M/19del mutant consistently shows increased activity compared with the T790M/L858R mutant, as measured by morphologic transformation, soft agar colony formation, and tumorigenicity assays (26). Yu et al. reported that the patients with 19del presented a longer PFS after first-line EGFR-TKI treatment (14.4 vs. 11.4 months, P=0.034) compared with those with L858R, although no statistically significant difference in OS was observed (27). Jackman et al. found that patients with 19del had a significantly longer OS (38 vs. 17 months, P=0.04) and improved PFS (24 vs. 10 months), although not independently significant in a multivariate analysis, compared with patients with L858R following treatment with gefitinib or erlotinib (28). However, Yu et al. reported that the PFS after EGFR-TKIs was similar for patients with 19del vs. L858R mutations (15 vs. 17 months, P=0.99) (8). Zhuo et al. also demonstrated that no significant difference was detected in PFS or OS between 19del and L858R groups after eliminating potential imbalanced factors, such as sex, age, histological type, clinical stage, brain metastases, mutation frequency and therapy line, through a propensity score matching method (25). In the present study, the results showed statistically significant difference neither in the proportion of 19del and L858R coexistence with acquired T790M nor in PFS1 between 19del group and L858R group. So at least for now, whether patients carrying 19del and L858R mutations exhibit different responsiveness to EGFR-TKIs and the potential mechanism for such difference remains controversial.
We demonstrated that higher mutant abundance of T790M was associated with a shorter PFS, which was supported by previous research that high mutant copy number of T790M (≥105 per mL of plasma) was correlated with shorter PFS (5.5 months vs. not achieved) and shorter OS (9.1 months vs. not achieved) (29). A high T790M mutant abundance might be indicative of a higher proportion of cells harboring the T790M mutation, leaving the tumor lesions more vulnerable to T790M-dependent resistance. Moreover, higher mutant abundance may also reflect higher tumor burden and higher aggressiveness.
We acknowledge several limitations to our study. First, the nature of retrospective and single-institution study might cause some statistical bias. Second, matched tissue and liquid biopsy results were not available for most of the patients; therefore, the concordance rate of T790M in gDNA and ctDNA could not be evaluated. Third, the follow-up time was not long enough to draw any conclusion on OS in these NSCLC patients.
Conclusions
In conclusion, ddPCR is a better choice of method to detect EGFR T790M mutation due to its higher sensitivity and ability to provide quantification of mutant abundance and copy number. Moreover, our study revealed that ctDNA was comparable with gDNA in detecting T790M by ddPCR, and patients with T790M mutation, especially those with low abundance, could benefit more from osimertinib.
Acknowledgments
Funding: We acknowledge financial support from the Natural Science Foundation of Guangdong Province (2017A030310192) and Fundamental Research Funds for the Central Universities (17ykpy84).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center (No. B2018-157-01), all patients provided signed informed consent, and the research was carried out in accordance with the Helsinki Declaration. The study outcomes will not affect the future management of the patients.
References
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or Chemotherapy for Non-Small-Cell Lung Cancer with Mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010;28:357-60. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR Mutation and Resistance of Non-Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and Common Secondary T790M Mutations in Epidermal Growth Factor Receptor-Mutant Lung Adenocarcinomas with Acquired Resistance to Kinase Inhibitors. Clin Cancer Res 2006;12:6494-501. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Sun JM, Ahn MJ, Choi YL, et al. Clinical implications of T790M mutation in patients with acquired resistance to EGFR tyrosine kinase inhibitors. Lung Cancer 2013;82:294-8. [Crossref] [PubMed]
- Ayoola A, Barochia A, Belani K, et al. Primary and Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-small Cell Lung Cancer: An Update. Cancer Invest 2012;30:433-46. [Crossref] [PubMed]
- Zhang BO, Xu CW, Shao Y, et al. Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. Exp Ther Med 2015;9:1383-8. [Crossref] [PubMed]
- Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med 2017;6:154-62. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [Crossref] [PubMed]
- Feng Q, Gai F, Sang Y, et al. A comparison of QuantStudio™ 3D Digital PCR and ARMS-PCR for measuring plasma EGFR T790M mutations of NSCLC patients. Cancer Manag Res 2018;10:115-21. [Crossref] [PubMed]
- Bai H, Mao L, Wang HS, et al. Epidermal Growth Factor Receptor Mutations in Plasma DNA Samples Predict Tumor Response in Chinese Patients With Stages IIIB to IV Non-Small-Cell Lung Cancer. J Clin Oncol 2009;27:2653-9. [Crossref] [PubMed]
- Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. [Crossref] [PubMed]
- Goto K, Ichinose Y, Ohe Y, et al. Epidermal Growth Factor Receptor Mutation Status in Circulating Free DNA in Serum. J Thorac Oncol 2012;7:115-21. [Crossref] [PubMed]
- Costa DB, Schumer ST, Tenen DG, et al. Differential Responses to Erlotinib in Epidermal Growth Factor Receptor (EGFR)-Mutated Lung Cancers With Acquired Resistance to Gefitinib Carrying the L747S or T790M Secondary Mutations. J Clin Oncol 2008;26:1182-4; author reply 1184-6. [Crossref] [PubMed]
- Bean J, Riely GJ, Balak M, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res 2008;14:7519-25. [Crossref] [PubMed]
- Chiba M, Togashi Y, Bannno E, et al. Efficacy of irreversible EGFR-TKIs for the uncommon secondary resistant EGFR mutations L747S, D761Y, and T854A. BMC Cancer 2017;17:281. [Crossref] [PubMed]
- Li W, Qiu T, Guo L, et al. Primary and acquired EGFR T790M-mutant NSCLC patients identified by routine mutation testing show different characteristics but may both respond to osimertinib treatment. Cancer Lett 2018;423:9-15. [Crossref] [PubMed]
- Liang H, Pan Z, Wang W, et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: a literature-based pooled analysis. J Thorac Dis 2018;10:2311-20. [Crossref] [PubMed]
- Chen LY, Molina-Vila MA, Ruan SY, et al. Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: A systematic review and meta-analysis. Lung Cancer 2016;94:46-53. [Crossref] [PubMed]
- Zhuo M, Zheng Q, Zhao J, et al. Survival difference between EGFR Del19 and L858R mutant advanced non-small cell lung cancer patients receiving gefitinib: a propensity score matching analysis. Chin J Cancer Res 2017;29:553-60. [Crossref] [PubMed]
- Godin-Heymann N, Bryant I, Rivera MN, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res 2007;67:7319-26. [Crossref] [PubMed]
- Yu JY, Yu SF, Wang SH, et al. Clinical outcomes of EGFR-TKI treatment and genetic heterogeneity in lung adenocarcinoma patients with EGFR mutations on exons 19 and 21. Chin J Cancer 2016;35:30. [Crossref] [PubMed]
- Jackman DM. Exon 19 Deletion Mutations of Epidermal Growth Factor Receptor Are Associated with Prolonged Survival in Non-Small Cell Lung Cancer Patients Treated with Gefitinib or Erlotinib. Clin Cancer Res 2006;12:3908-14. [Crossref] [PubMed]
- Li JY, Ho JC, Wong KH. T790M mutant copy number quantified via ddPCR predicts outcome after osimertinib treatment in lung cancer. Oncotarget 2018;9:27929-39. [Crossref] [PubMed]


