
The prognostic value of tumor mutation burden in EGFR-mutant advanced lung adenocarcinoma, an analysis based on cBioPortal data base
Introduction
In the era of precision medicine, more and more genetic information detected by next generation sequence (NGS) is accumulating and changing the treatment of lung cancer (1-3). In addition to traditional targets such as EGFR, anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1 (ROS1), MET, HER-2, BRAF, etc. Large test panels such as MSK-IMPACT (4), FMI (5) and Burning Rock (6) can provide more information, especially TMB, which can be only presented in the era of NGS. How to excavate the meaning of these massive data and apply it to daily treatment of lung cancer is of great practical significance.
TMB is a novel biomarker of promising predict value in prediction of immune checkpoint inhibitors (ICPis) in lung cancer. Many studies showed that in patients with high TMB, immunotherapy had a better effect (7-13). However, a considerable portion of non-small cell lung cancer (NSCLC) patients have EGFR mutations, especially in the East Asian population. The distribution of TMB in EGFR-mutant NSCLC patients and the prognostic and predictive value of TMB in EGFR-mutant NSCLC patients have not been well established in the first few years of clinical application of TMB.
Recently, TMB was reported to be negatively associated with clinical outcomes in metastatic EGFR-mutant lung cancer patients treated with EGFR tyrosine kinase inhibitor (TKI), including progression-free survival (PFS) and overall survival (OS) (14). However, in early stage resected NSCLC patients, the prognostic value of TMB was reverse as compared with late stage (15). Meanwhile, this study was based on Caucasian population, which have an obvious different EGFR-mutant rate compared with Chinese population. Further investigate TMB in EGFR-mutant NSCLC patient is meaningful.
Our previous study revealed that TP53 had important prognostic value in EGFR-mutant NSCLC patients (16). And TP53 mutation rates were diverse in patients with different TMB level (14). Whether TP53 and TMB together could predict OS more accurately in EGFR-mutant NSCLC patients is still unknown.
Our study was conducted to reveal the distribution of TMB in different EGFR-mutant groups, especially in Chinese population to discuss the prognostic value of TMB in EGFR-mutant advanced lung adenocarcinoma (LUAD), as well as the correlation of TMB and other co-mutations such as TP53 in this group of patients.
Methods
Information regarding TMB, EGFR alterations and patients’ survival time in NSCLC was downloaded from The Cancer Genome Atlas (TCGA) database, an open access database that is available at http://www.cbioportal.org (17,18). Using the TCGA database, the test data of MSKCC-IMPACT (19) including 1,668 cases of LUAD patients were retrieved. Patients with EGFR mutation and survival data were analyzed to observe the prognostic value of TMB. MSK-IMPACT is an FDA cleared NGS test in which tumor and normal DNA undergo targeted hybridization capture and deep-coverage NGS to detect somatic mutations, copy number changes, and select gene fusions in a custom gene panel of 341 (version 1), 410 (version 2), or 468 (version 3) genes (4). TMB was defined as the total number of non-synonymous single nucleotide or insertion/deletion mutations divided by the coding region captured in each panel (341 genes, 0.98 Mb; 410 genes, 1.06 Mb; 468 genes, 1.22 Mb). TMB value was presented in the form of mutation number per megabase pairs.
Another cohort of Chinese advanced LUAD patient was from Burning Rock company. We use this cohort to further observe the distribution of TMB in advanced LUAD. All these patients were sequenced by two separated gene panels, which were presented as a custom gene panel of 295 or 520 genes. The detailed NGS library and sequencing protocol preparation was performed as previously described (6). TMB was defined as the number of somatic mutations excluding copy number variations (CNVs), fusions and large genome rearrangement (LGR) per megabase of genome examined. To be more specific, the mutations counted included missense, synonymous, frameshift, splice site and indel mutations whereas the genomic regions examined included all coding sequences extending 20 bp into the introns. The kinase domains of EGFR and ALK genes were excluded for TMB calculation. Thus, the total examined regions were 0.98 Mb for 295 panel and 1.26 Mb for 520 panel. TMB value was also presented in the form of mutation number per megabase pairs.
The date from TCGA database do not require ethical approval. This study was approved by the Committee on Ethics of Changzheng Hospital, Shanghai (No. 2017SL016).
Statistical methods
All the analyses were performed by SPSS 20.0. The continuous variables were described by median and range. The difference between the two groups was analyzed by Mann-Whitney U test. Comparison between three or more groups was analyzed by Kruskal-Wallis test with Dunn’s post-test. Survival time was analyzed by Kaplan-Meier curves, where P value was determined by Log-rank test. Hazard’s ratio and its 95% CI were determined by univariate and multivariate Cox proportional hazards model. All reported P values were two-tailed and P<0.05 was considered statistically significant.
Results
The distribution of TMB in advanced LUAD patients
In the MSKCC-IMPACT study, there were 1,668 cases with advanced LUAD. Of the 1,668 cases, 1,258 cases were EGFR wild type patients, and 410 cases were with EGFR mutations. Among the 410 mutant patients, 172 cases had exon 19 deletions (19del), 131 cases had L858R, and 107 cases demonstrated other types of mutations. The median TMB values of EGFR wild-type, non-sensitive EGFR mutations, 19del and L858R were 6.12, 5.66, 3.77 and 4.72, respectively (Table 1). Difference between wild-type and EGFR 19del was significant (P<0.001, Figure 1A).
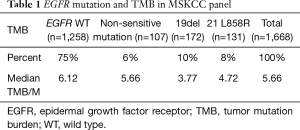
Full table
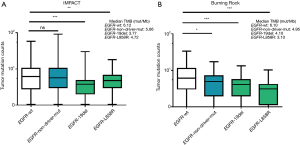
We used another cohort of Chinese data from Burning Rock Company to further verify the distribution of TMB in advanced LUAD. A total of 292 advanced LUAD patients were included. In these patients, 168 cases were EGFR wild-type patients, and 124 cases had EGFR mutations. Among the 124 EGFR-mutant patients, 44 cases had 19del, 28 cases had L858R, and 52 cases had other types of mutations. The median TMB values of EGFR wild-type, non-sensitive EGFR mutations, 19del and L858R were 6.10, 4.95, 4.10 and 3.10 respectively (Table 2). Differences between wild-type and each mutant group were significant (EGFR-19del vs. EGFR wild-type, P<0.001; EGFR-L858R vs. EGFR wild-type, P<0.001; other types of EGFR mutation vs. EGFR wild-type, P<0.05, Figure 1B).

Full table
Prognostic significance of TMB in advanced LUAD patients with EGFR sensitive mutations
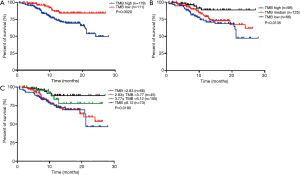
In MSKCC-IMPACT cohort, 289 patients with both survival data and EGFR sensitive mutations (19del or L858R) data were analyzed to further reveal the prognostic value of TMB in EGFR-mutant advanced LUAD. In this specific population, the median TMB was 3.77. When we divided these patients into TMB high group (TMB ≥3.77) and TMB low group (TMB <3.77), the median OS time was 24.03 months and not reached (P=0.0020, Figure 2A). When we divided these patients into 3 groups by tertile (TMB high: TMB ≥5.10, TMB median: 2.83≤ TMB <5.10, TMB low: TMB <2.83), the median OS time was 21.27 months, not reached and not reached (P=0.0135, Figure 2B). Although median survival times were not reached, but the survival time was significantly longer in patients with low or median TMB than high TMB. However, the survival curve of high TMB and median TMB were overlapping. If these patients were divided into 4 groups by quartile (TMB ≥6.12, 3.77≤ TMB <6.12, 2.83≤ TMB <3.77, TMB <2.83), the median OS time was 21.27 months, not reached, not reached and not reached (P=0.0180, Figure 2C). Similarly, although median survival times were not reached, but the survival time was significantly longer in patients with low or intermediate TMB than high TMB. The survival curve of the top two TMB groups were overlapping. As above, we use median TMB as cutoff value to divide TMB into high or low groups for further analyses.
We further carried out multivariate analysis to determine the prognostic value of TMB. Due to limited information by the MSKCC-IMPACT database, only gender and smoking status were obtained. However, all the sequencing data was from advanced NSCLC patients, so tumor staging of these patients was almost the same. As our former article had mentioned, TP53, EGFR, SMARCA4, STK11, gender and smoking status were statistically significant factors in univariate analysis in advanced NSCLC (16). Hence, we further performed univariate and cox regression analysis.
In univariate analysis, only TP53, TMB, STK11 and gender was statistically significant, these 4 factors were included in multivariate analysis, and all these factors was statistically significant. We confirmed that TMB was an independent prognostic factor in EGFR-mutant advanced LUAD (Table 3).
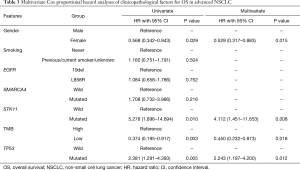
Full table
Correlation of TP53 gene mutation and high TMB in advanced EGFR-mutant LUAD
As TP53 was the most common mutated gene in NSCLC, so we observed the correlation of TP53 and TMB in advanced EGFR-mutant LUAD. We found that in TMB high group, TP53 mutation rate was 76.4% (136 in 178), in TMB low group, TP53 mutation rate was 47.7% (53 in 111), the difference was statistically significant (P<0.001).
Prognostic value of TMB and TP53 in advanced EGFR-mutant LUAD
In our previous study, we found that TP53 was an important prognostic factor in advanced LUAD, and the different mutation site of TP53 has different prognostic value (16). At the same time, TP53 mutation rate was higher in TMB high group, so we were wondering whether TMB combined with TP53 may be a better prognostic factor in advanced EGFR-mutant LUAD. Thus, we made further statistically analysis.
When we consider TMB and TP53 mutation together, we can classify these patients into four groups. Group A, TMB high and TP53 wild type. Group B, TMB high and TP53 mutant type. Group C, TMB low and TP53 wild type. Group D, TMB low and TP53 mutant type. The OS disparity of them was statistically significant (P=0.0009). However, the survival curves of group A, group C and group D were overlapping, each of the 3 groups has better OS than group B (Figure 3A). When we integrate group A, group C and group D together, we can further classify these patients into two groups. Group A, TMB high and TP53 mutant type. Group B, non-TMB high and TP53 mutant type. The survival curve of these two groups was separated and statistically significant (P<0.0001, Figure 3B). It is likely that TMB high and TP53 mutation were both important for the prediction of OS in advanced EGFR-mutant LUAD. The former viewpoint of judge the prognosis only by TP53 or TMB may mis-divide some patients with better prognosis into worse prognosis group.

Discussion
With the wide application of targeted drugs, the treatment of lung cancer has entered the era of precision treatment. As a therapeutic target with the highest mutation rate in LUAD, the total mutation rate of EGFR in east Asian LUAD was as high as 50.2% (20), and its expression was associated with poor prognosis (21). In addition, the drug-sensitive mutation rate of EGFR is high. 19del accounted for 45%, exon 21 L858R mutation accounted for 40%, and the remaining 10% of mutations involved exons 18 and 20 (21). Therefore, EGFR is the most important target in lung cancer therapy. The I subclass of the receptor tyrosine kinase (RTK) superfamily consists of ERBB or epidermal growth factor (EGF) receptors and includes four members: EGFR/ERBB1, ERBB2, ERBB3, and ERBB4 (22,23). All members have an extracellular ligand binding region, a single transmembrane region and a cytoplasmic tyrosine kinase domain (22,24). The extracellular partial binding of EGFR ligand and receptor successfully activates the active dimer, whose autophosphorylation activates the RAS-RAF-MEK-mitogen-activated protein kinase (MAPK) pathway and phosphatidylinositol 3-kinase (PI3K)-Akt pathways (25). MAPK signaling pathway is mainly involved in the regulation of cell proliferation, differentiation, survival and invasion. PI3K/Akt pathway controls various important cell functions including glucose metabolism, cell proliferation and survival (26).
With the wide application of NGS, TMB has been paid more and more attention to the role of cancer screening, surveillance, and therapy (27). The definition of TMB is the total number of mutations per coding area of a tumor genome (28). It was also called as tumor mutation load (TML) or tumor mutational burden (8,9). TMB showed differences in different races, and the difference in TMB in different races was associated with the incidence of cancer (29). Somatic mutations in tumor cells may produce neoantigens, and the recognition of neoantigens by T-cells seems to be important for the activity of programmed cell death protein 1 (PD-1) pathway inhibitors (9). Several studies have suggested that tumors harboring high levels of somatic mutations might be highly sensitive to ICPis (12,30). TMB is not only related to immunotherapy, but also can predict the efficacy of different chemotherapy regimens. It was found that in low TMB group of colorectal cancer patients, PFS was longer in regimens containing irinotecan than oxaliplatin (11.9 vs. 6.5 months, P<0.001) (31). In addition to the predictive value, TMB was thought to be associated with cancer prognosis, such as head and neck squamous cancer, lung cancer and so on (29).
NSCLC was found to have higher TMB than many other cancer types except melanoma, where ICPis was first used (10). In NSCLC, TMB was also an important factor in the predictive and prognostic aspects. Many studies have found that TMB was positively related to the effect of ICPis (9,32). Rizvi et al. demonstrated that the median number of nonsynonymous mutations was higher in patients with durable clinical benefit (DCB) (partial or stable response lasting for >6 months) vs. those with no durable benefit (NDB) (12). McGranahan found that sensitivity to PD-1 and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) blockade in patients with advanced NSCLC was enhanced in tumors enriched for clonal neoantigens. At the same time, a high neoantigen burden, which was defined as the upper quartile of neoantigen load, was associated with significantly longer OS in LUAD (33). In NSCLC patients, without considering EGFR mutation, a 51 gene panel sequencing indicated that the prognosis of the patients with high TMB was inferior to those with low TMB (34). In Chinese population, the results also demonstrated that greater burden of genetic alterations was associated with worse OS rate, and multivariable analysis demonstrated that the mutation burden was an independent prognostic factor for the patients (34). This disparity of OS in LUAD of high TMB may be caused by the administration of ICPis in the study population.
In EGFR-mutant advanced NSCLC patients, results of ICPis remained controversial. Although the preclinical results indicated that programmed cell death-ligand 1 (PD-L1) expression was higher in EGFR-TKI resistant cells harboring EGFR-T790M mutation and suggested that PD-1/PD-L1 blockade may be a promising approach in pre-treated EGFR-mutant advanced NSCLC patients (35). But these results were not replicated in the clinical trials (36,37). Furthermore, a study in Korea found that lung cancer patients with T790M mutation after using EGFR-TKI had lower TMB, confirming the lower efficacy on ICPis from the point of mechanism (38). Recently, a meta-analysis suggested that the use of ICPis in pre-treated EGFR-mutant advanced NSCLC patients had similar OS but worse PFS when compared with docetaxel, whereas both OS and PFS were in favor of ICPis in the wild-type EGFR population (39). In consideration of the positive predictive value of TMB to the effect of ICPis, TMB in EGFR-mutant advanced NSCLC patients may be inferior to those with wild-type EGFR, so ICPis is unlikely to be effective in EGFR-mutant advanced NSCLC patients. In patients with EGFR mutations, we already know that tumor burden was associated with the prognosis of patient (40-42). Recently, TMB was proved to be an adverse prognostic factor in EGFR-mutant late stage NSCLC patients (14), but the prognostic value was opposite in resected early stage NSCLC patients (15). Meanwhile, EGFR-mutant late stage NSCLC patients with higher TMB benefitted less in administration of EGFR-TKIs (14). However, the diversity of TMB in EGFR-mutant Chinese NSCLC patients is still unknown, which has a totally different EGFR mutation rate compared with Caucasia population. Furthermore, the prognostic value of TMB and TP53 together was not well demonstrated.
Our study indicated that in late stage Chinese LUAD patients, TMB was lower in patient with EGFR-mutant group than EGFR wild group. We verified that TMB was a negative prognostic biomarker of OS in EGFR-mutant LUAD patients. Furthermore, we first present a novel viewpoint that we should use TMB and TP53 mutation together to judge the prognosis of EGFR-mutant LUAD patients more accurately.
However, OS data in our study was downloaded from TCGA database, several clinical details such as treatment regimens and biopsy time could not be acquired. On the other hand, the majority of EGFR-mutant Chinese patients was being treated with EGFR-TKI, so the predictive value of TMB to the efficacy of EGFR-TKI could not been presented in this study. Further studies should be conducted to deeply understand the meaning of TMB as well as other co-mutations in EGFR-mutant LUAD patients.
Conclusions
In late stage LUAD patients, TMB was lower in patients with EGFR-mutant group than EGFR wild group. TMB was a negative prognostic biomarker of OS in EGFR-mutant LUAD patients, especially when TP53 was mutated together. The prognostic value of TMB as well as other co-mutations in late stage EGFR-mutant LUAD patient needs to be further investigated.
Acknowledgments
We thank Medsci (www.medsci.cn) for its linguistic assistance during the preparation of this manuscript. We thank Burning Rock Company for their data.
Funding: This work received the support of Shanghai Municipal Science and Technology Commission Foundation (grant number 17511103403).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Committee on Ethics of Changzheng Hospital, Shanghai (No. 2017SL016).
References
- Rozenblum AB, Ilouze M, Dudnik E, et al. Clinical Impact of Hybrid Capture-Based Next-Generation Sequencing on Changes in Treatment Decisions in Lung Cancer. J Thorac Oncol 2017;12:258-68. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018;6:681-90. [Crossref] [PubMed]
- Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Shien K, Papadimitrakopoulou VA, Wistuba II. Predictive biomarkers of response to PD-1/PD-L1 immune checkpoint inhibitors in non-small cell lung cancer. Lung Cancer 2016;99:79-87. [Crossref] [PubMed]
- Colli LM, Machiela MJ, Myers TA, et al. Burden of Nonsynonymous Mutations among TCGA Cancers and Candidate Immune Checkpoint Inhibitor Responses. Cancer Res 2016;76:3767-72. [Crossref] [PubMed]
- Oberndorfer F, Mullauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol 2018;30:69-76. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Campesato LF, Barroso-Sousa R, Jimenez L, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 2015;6:34221-7. [Crossref] [PubMed]
- Offin M, Rizvi H, Tenet M, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res 2019;25:1063-9. [Crossref] [PubMed]
- Devarakonda S, Rotolo F, Tsao MS, et al. Tumor Mutation Burden as a Biomarker in Resected Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:2995-3006. [Crossref] [PubMed]
- Jiao XD, Qin BD, You P, et al. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018;123:70-5. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. [Crossref] [PubMed]
- Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med 2005;353:172-87. [Crossref] [PubMed]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341-54. [Crossref] [PubMed]
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 2009;21:177-84. [Crossref] [PubMed]
- Shostak K, Chariot A. EGFR and NF-kappaB: partners in cancer. Trends Mol Med 2015;21:385-93. [Crossref] [PubMed]
- Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160-74. [Crossref] [PubMed]
- Quatrale AE, Porcelli L, Silvestris N, et al. EGFR tyrosine kinases inhibitors in cancer treatment: in vitro and in vivo evidence. Front Biosci (Landmark Ed) 2011;16:1962-72. [Crossref] [PubMed]
- Campbell BB, Light N, Fabrizio D, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017;171:1042-56.e10. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Zhang W, Edwards A, Flemington EK, et al. Racial disparities in patient survival and tumor mutation burden, and the association between tumor mutation burden and cancer incidence rate. Sci Rep 2017;7:13639. [Crossref] [PubMed]
- Champiat S, Ferte C, Lebel-Binay S, et al. Exomics and immunogenics: Bridging mutational load and immune checkpoints efficacy. Oncoimmunology 2014;3:e27817. [Crossref] [PubMed]
- Pai SG, Carneiro BA, Chae YK, et al. Correlation of tumor mutational burden and treatment outcomes in patients with colorectal cancer. J Gastrointest Oncol 2017;8:858-66. [Crossref] [PubMed]
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Xiao D, Pan H, Li F, et al. Analysis of ultra-deep targeted sequencing reveals mutation burden is associated with gender and clinical outcome in lung adenocarcinoma. Oncotarget 2016;7:22857-64. [Crossref] [PubMed]
- Chen N, Fang W, Zhan J, et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J Thorac Oncol 2015;10:910-23. [Crossref] [PubMed]
- Dong ZY, Zhang JT, Liu SY, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 2017;6:e1356145. [Crossref] [PubMed]
- Lee CK, Man J, Lord S, et al. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol 2017;12:403-7. [Crossref] [PubMed]
- Lee CK, Kim S, Lee JS, et al. Next-generation sequencing reveals novel resistance mechanisms and molecular heterogeneity in EGFR-mutant non-small cell lung cancer with acquired resistance to EGFR-TKIs. Lung Cancer 2017;113:106-14. [Crossref] [PubMed]
- Soo RA, Lim SM, Syn NL, et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non-small cell lung cancer: Current controversies and future directions. Lung Cancer 2018;115:12-20. [Crossref] [PubMed]
- Cha YK, Lee HY, Ahn MJ, et al. Survival outcome assessed according to tumor burden and progression patterns in patients with epidermal growth factor receptor mutant lung adenocarcinoma undergoing epidermal growth factor receptor tyrosine kinase inhibitor therapy. Clin Lung Cancer 2015;16:228-36. [Crossref] [PubMed]
- Park JH, Kim TM, Keam B, et al. Tumor burden is predictive of survival in patients with non-small-cell lung cancer and with activating epidermal growth factor receptor mutations who receive gefitinib. Clin Lung Cancer 2013;14:383-9. [Crossref] [PubMed]
- Zhao B, Oxnard GR, Moskowitz CS, et al. A pilot study of volume measurement as a method of tumor response evaluation to aid biomarker development. Clin Cancer Res 2010;16:4647-53. [Crossref] [PubMed]


