
Circ_0058063 upregulates GLUT1 expression and promotes glucose-uptake in esophageal squamous-cell carcinomas
Introduction
Esophageal cancer affects more than 450,000 new cases very year (1). As a major subtype of esophageal cancer, esophageal squamous-cell carcinomas (ESCC) is responsible for about 90% of all cases (1). Incidence and mortality rates of ESCC vary a lot across the world, and the highest incidence has been observed in Asia, which is possibly due to the economic gains and dietary structure (2,3). In western countries, ESCC is mainly caused by cigarette smoking (1,4). Both incidence and mortality rates of ESCC decline continuously in recent years (5). However, in China, ESCC is still a major cause of cancer-related mortalities (2,3). Therefore, novel therapeutic approaches are needed.
Circular RNA, also known as cirRNA, is a group of circular form single-stranded RNA with limited or no protein coding capacity (6). Different from other genes, expression of cirRNA is usually cell-type specific, which supports their roles in specific cellular processes (7). CirRNA also have critical roles in cancer biology, such as by sponging tumor suppressive or oncogenic miRNAs (8,9). However, the functions of most cirRNAs in cancer biology remain unknown. In a recent study, Sun et al. reported an oncogenic cirRNA named Circ_0058063 as an oncogene in bladder cancer (10). We performed a microarray analysis and found that Circ_0058063 was correlated with GLUT1, which is usually upregulated in cancer to promote glucose metabolism (11). Inhibition of glucose metabolism has been proven as a promising target for cancer therapies by inhibiting cancer cell growth and metastasis (11). Therefore, in-depth analysis of the mechanisms that regulate the expression of GLUT1 may provide novel insights to the development of anti-cancer therapies. This study aimed to investigate the interaction between Circ_0058063 and GLUT1 in ESCC.
Methods
ESCC patients
This study passed the review board of Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. A total of 60 ESCC patients were selected from the 102 ESCC cases admitted to Tianjin Medical University Cancer Institute and Hospital between March 2015 and March 2019. Inclusion criteria: (I) newly diagnosed ESCC cases; (II) confirmed by histopathological biopsy; (III) therapies were not initiated. Exclusion criteria: (I) recurrent ESCC; (II) history of malignancies; (III) other clinical disorders were combined; (IV) therapies were performed. Based on AJCC staging system and clinical findings, the 60 ESCC patients included 9, 14, 19 and 18 cases at clinical stage I–IV, respectively. All patients were informed with the experimental principle, and informed consent was provided by all of them.
ESCC tissue samples and cells
MRI-guided biopsy was performed on all the 60 ESCC patients to collect both non-tumor and ESCC tissue samples. The weight of samples ranged from 0.012 to 0.023 g. All tissue samples were checked by histopathological examinations and all ESCC tissues contained more than 90% cancer cells and all non-tumor samples contained less than 1% cancer cells.
Human ESCC cell line KYSE510 from ATCC (USA) was used. Cells were cultivated in a mixture of 90% RPMI-1640 medium and 10% FBS. Cells were cultivated under the conditions of 37 °C, 5% CO2 and 95% humidity.
Cell transfection
KYSE510 cells were harvested at confluence of 70–80% to perform cell transfections. Vectors expressing Circ_0058063 and GLUT1 were constructed by RIBOBIO (Guangzhou, China). Circ_0058063 siRNA and negative control (NC) siRNA were also from RIBOBIO. All transfections were performed using lipofectamine 2000 (RIBOBIO) to transfect 40 nM siRNA (NC siRNA as NC group) and 10 nM vector (empty vector as NC group) into 106 cells. Control (C) cells for all transfections were untransfected cells. Following studies were performed at 24 h post-transfection.
Gene expression analysis by quantitative polymerase chain reaction (qPCR)
KYSE510 cells were harvested at 24 post-transfection and were counted. Total RNAs in 105 cells and 0.01 g tissue samples were extracted using Trizol reagent (Invitrogen, USA). All RNA samples were digested for 2 h with DNase I at 24 °C to remove genomic DNA. Following that, reverse transcriptions were performed using RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). To measure the relative expression levels of Circ_0058063 and GLUT1, all qPCR mixtures were prepared using DyNAmo Flash SYBR Green qPCR Kit (Thermo Fisher Scientific) with GAPDH as the endogenous control. All data were processed using 2–ΔΔCT method, and each experiment was performed in 3 replicates.
Glucose uptake analysis
KYSE510 cells were harvested at 24 post-transfection and were counted. 105 cells were incubated with 2 mL glucose-free RPMI-1640 medium in a well of a 6-well cell culture plate for 16 under aforementioned conditions, followed by incubation in high-glucose RPMI-1640 medium under the same cell culture conditions for further 24 h. Finally, intracellular glucose levels were measured using a fluorescence-based glucose assay kit (BioVision, Milpitas, CA, USA).
Cell proliferation assay
KYSE510 cells were harvested at 24 post-transfection and were counted. 5×104 cells were mixed with 1 mL mixture of 90% RPMI-1640 medium and 10% FBS. Cells were then cultivated in a 96-well cell culture plate under aforementioned conditions, and 10 µL CCK-8 solution (Sigma-Aldrich, St. Louis, MO, USA) was added into each well at 3 h before the termination of cell culture. After cell culture was stopped, each well was added with 10 µL DMSO, followed by measurement of optical density (OD) values at 450 nm.
Western-blot
KYSE510 cells were harvested at 24 post-transfection and were counted. Total proteins were extracted from 105 cells using RIPA solution (RIBOBIO). To denature proteins, protein samples were incubated with boiling water for 10 min. After that, electrophoresis (10% SDS-PAGE gel) was performed to separate proteins, followed by transferring proteins to PVDF membranes. To block membranes, all PVDF membranes were incubated with 5% non-fat milk for 1 h at room temperature. After that, rabbit anti-GAPDH antibody (1:1,000, ab9485, Abcam, Cambridge, UK) and anti-GLUT-1 (1:1,500, ab15309, Abcam) primary antibodies were first used to incubate the membranes for 12 h at 4 °C. After that, IgG-HRP secondary antibody (goat anti-rabbit, 1:1,500, MBS435036, MyBioSource, San Diego, CA, USA) was used to incubate with the membranes for further 2 h. Finally, membranes were incubated with ECLTM Blocking Agent (GE Healthcare, Chicago, IL, USA) for 5 min at room temperature, and data were processed using Image J V1.34 software.
Statistical analysis
Experiments were performed in 3 biological replicates and mean values were calculated. Linear regression was used for correlation analyses. Paired t-test and analysis of variance (ANOVA) (one-way) in combination with Tukey test were used to explore differences between two types of tissues (ESCC vs. non-tumor) and among multiple cell groups, respectively. P<0.05 was statistically significant.
Results
Circ_0058063 and GLUT1 mRNA were both upregulated in ESCC
Measurement of expression levels of Circ_0058063 and GLUT1 mRNA was performed by qPCR and expression data were compared by performing paired t-test. It can be observed that expression levels of Circ_0058063 in ESCC tissues were significantly higher than those in non-tumor tissues (Figure 1A, P<0.05). Moreover, GLUT1 mRNA levels were also significantly higher in ESCC tissues than in non-tumor tissues (Figure 1B, P<0.05).

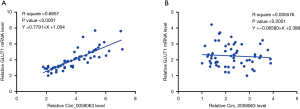
Circ_0058063 and GLUT1 mRNA were correlated in ESCC
Correlations between Circ_0058063 and GLUT1 mRNA were analyzed by linear regression. It can be observed that expression levels of Circ_0058063 were significantly and positively correlated with the expression levels of GLUT1 mRNA in ESCC tissues (Figure 2A). However, the correlation between Circ_0058063 and GLUT1 mRNA was not significant in non-tumor tissues (Figure 2B).
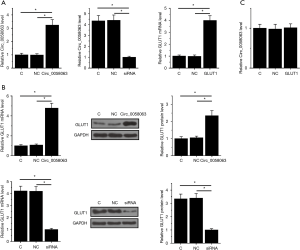
Circ_0058063 positively regulated GLUT1 in KYSE510 cells
Circ_0058063 and GLUT1 expression vectors as well as Circ_0058063 siRNAs were transfected into KYSE510 cells. Comparing to NC and C groups, qPCR data showed that expression of Circ_0058063 and GLUT1 was significantly altered at 24 h post-transfection (Figure 3A, P<0.05). Moreover, Circ_0058063 overexpression resulted in the upregulated, while Circ_0058063 knockdown resulted in downregulated GLUT1 at both mRNA and protein levels (Figure 3B, P<0.05). Moreover, GLUT1 overexpression failed to affect Circ_0058063 expression (Figure 3C).
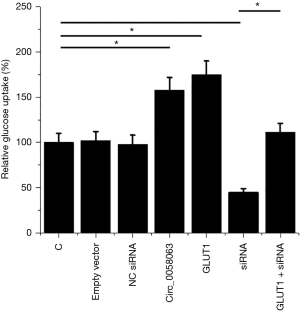
Circ_0058063 promoted glucose uptake in KYSE510 cells through GLUT1
Glucose uptake assay was performed to investigate the effects of Circ_0058063 and GLUT1 overexpression as well as Circ_0058063 siRNA silencing on glucose uptake in KYSE510 cells. Comparing to C and NC groups, Circ_0058063 and GLUT1 overexpression promoted glucose uptake in KYSE510 cells, while Circ_0058063 siRNA silencing inhibited glucose uptake. Moreover, GLUT1 overexpression attenuated the effects of Circ_0058063 siRNA silencing (Figure 4, P<0.05).
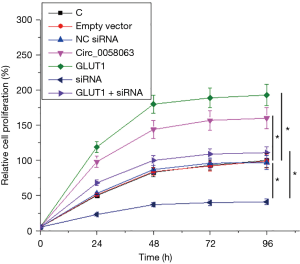
Circ_0058063 promoted KYSE510 cell proliferation through GLUT1
Cell proliferation assay was performed to investigate the effects of Circ_0058063 and GLUT1 overexpression as well as Circ_0058063 siRNA silencing on the proliferation of KYSE510 cells. Comparing to C and NC groups, Circ_0058063 and GLUT1 overexpression promoted led to increased, while Circ_0058063 siRNA silencing led to decreased rate of KYSE510 cell proliferation. Moreover, GLUT1 overexpression attenuated the effects of Circ_0058063 siRNA silencing (Figure 5, P<0.05).
Discussion
This study mainly investigated the functions of Circ_0058063 in ESCC. We found that Circ_0058063 was upregulated in ESCC and Circ_0058063 may regulate glucose uptake in ESCC cells by positively regulating the expression of GLUT1, which has critical roles in glucose uptake (12).
The function of Circ_0058063 has only been investigated in bladder cancer (10). In bladder cancer, Circ_005806 was overexpressed and overexpression of Circ_005806 sponged miR-145-5p to upregulate CDK6, which in turn promoted the cell cycle progression and cell proliferation of bladder cancer cells (10). This study is the first to report the upregulation of Circ_0058063 in ESCC. We also showed the increased ESCC cell proliferation rate after Circ_0058063 overexpression. Therefore, Circ_0058063 plays oncogenic role in ESCC.
CirRNAs have critical functions in many aspects of cancer biology, such as epithelial-to-mesenchymal transition (13). Comparing to normal cells, energy metabolism is accelerated in cancer cells (14). Therefore, blocking of cancer metabolism, such as reducing glucose uptake, is a potential therapeutic approach for cancer treatment (15). Based on our knowledge, the interactions between cirRNAs and energy metabolism remain hardly known. This study showed that Circ_0058063 can positively regulate the glucose uptake in ESCC cells. Therefore, downregulation of Circ_0058063 may serve as a potential target to inhibit glucose consumption in ESCC cells.
However, the mechanism of the positive regulation of GLUT1 by Circ_0058063 is still unknown. It is known that Circ_0058063 can upregulate CDK6 to promote the proliferation of bladder cancer (10). It is known that the downregulation of CDK6 can lead to the downregulation of GLUT1 to inhibit the proliferation of ESCC cells (16). It is also known that CDK6 is upregulated in ESCC (17). Therefore, the altered expression of CDK6 may mediate the interaction between GLUT1 and Circ_0058063. It is known that may sponge tumor suppressive or oncogenic miRNAs to upregulate their downstream targets (8,9). Therefore, our future studies will explore the involvement of miRNAs in this process.
It has been well established that the inhibition of glucose metabolism may contribute to the treatment of cancers by reducing energy supply to cancer cells, thereby inhibiting multiple cancer cell behaviors, such as proliferation, migration and invasion (11). In this study we proved that Circ_0058063 is an upstream positive regulator of GLUT1 expression and it also promotes ESCC cell proliferation. Therefore, Circ_0058063 may serve as a potential therapeutic target for ESCC. However, clinical trials are needed to analyze clinical values of Circ_0058063 for ESCC treatment.
In conclusion, Circ_0058063 was upregulated in ESCC and promotes the proliferation of ESCC cell by upregulating GLUT1 to increase glucose uptake.
Acknowledgments
Funding: This work was supported by North Sichuan Medical College Scientific Research and Development Projects (No. CBY18-A-YB24); National Science Foundation of China (No. 81772619).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by National Human Genetic Resources Sharing Service Platform (No. 2005DKA21300).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology 2018;154:360-73. [Crossref] [PubMed]
- Zhang Y, Zheng T, Zhang W. Report of cancer incidence and mortality in China, 2012. Adv Mod Oncol Res 2018;4:1-7.
- Chen WQ, Li H, Sun KX, et al. Report of cancer incidence and mortality in China, 2014. Zhonghua Zhong Liu Za Zhi 2018;40:5-13. [PubMed]
- Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg 2018;41:210-5. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [Crossref] [PubMed]
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett 2017;396:138-44. [Crossref] [PubMed]
- Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer 2018;25:1-7. [Crossref] [PubMed]
- Sun M, Zhao W, Chen Z, et al. Circ_0058063 regulates CDK6 to promote bladder cancer progression by sponging miR-145-5p. J Cell Physiol 2019;234:4812-24. [Crossref] [PubMed]
- Yamamoto T, Seino Y, Fukumoto H, et al. Over-expression of facilitative glucose transporter genes in human cancer. Biochem Biophys Res Commun 1990;170:223-30. [Crossref] [PubMed]
- Meireles P, Sales-Dias J, Andrade CM, et al. GLUT1-mediated glucose uptake plays a crucial role during Plasmodium hepatic infection. Cell Microbiol 2017. [Crossref] [PubMed]
- Joseph NA, Chiou SH, Lung Z, et al. The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J Hematol Oncol 2018;11:74. [Crossref] [PubMed]
- Kim SY. Cancer energy metabolism: shutting power off cancer factory. Biomol Ther (Seoul) 2018;26:39-44. [Crossref] [PubMed]
- Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med 2012;209:211-5. [Crossref] [PubMed]
- Sawayama H, Ogata Y, Ishimoto T, et al. Glucose transporter 1 regulates the proliferation and cisplatin sensitivity of esophageal cancer. Cancer Sci 2019;110:1705-14. [Crossref] [PubMed]
- Li F, Xu Y, Liu B, et al. YAP1-Mediated CDK6 Activation Confers Radiation Resistance in Esophageal Cancer - Rationale for the Combination of YAP1 and CDK4/6 Inhibitors in Esophageal Cancer. Clin Cancer Res. 2019;25:2264-77. [Crossref] [PubMed]


