
Genomic alterations in thymoma—molecular pathogenesis?
Introduction
Thymomas and thymic carcinomas (TCs) are rare neoplasms derived from thymic epithelial cells. Thymomas are classified as A, AB, B1, B2 and B3 subtypes dependent on cell morphology and relative proportion of neoplastic epithelial cells and admixed non-neoplastic T-lymphocytes (1). Types A and AB thymomas are indolent tumors. On the other hand, B1, B2 and B3 thymomas exhibit an increasing propensity for intrathoracic spread. However, they usually do not seed hematogenous or lymphatic metastases. TCs, in contrast, are aggressive tumors, mostly squamous cell carcinomas, with a tendency for lymphatic and hematogenous dissemination. Thymic neuroendocrine tumors (NETs) comprise only about 5% of thymic epithelial tumors (TETs). They are morphologically classified similarly to lung NETs into carcinoid (typical and atypical), large cell neuroendocrine carcinoma and small cell carcinoma (1).
The causes for the development of TETs are unknown. There are no identified exogenous factors initiating or promoting the development of TETs. Furthermore, no family clustering has been observed, except for an increased prevalence among Asians/Pacific Islanders (2). The TETs are predominantly a disease of adulthood and exceedingly uncommon in children and adolescents (3). The incidence peaks in the seventh decade of life. This pattern is similar to many other tumors and most likely reflects the accumulation of genetic damage with age.
Thymomas may manifest concurrently with autoimmune diseases, most frequently myasthenia gravis (MG). Ten to 20% of MG patients develop a thymoma and about 30% of thymomas are associated with MG (4,5). Other thymic pathologies associated with MG are follicular thymitis and, in rare cases, thymolipoma (5). However, the thymus frequently appears histologically normal for age. TCs, in contrast, are not linked with MG or other specific autoimmune diseases, although paraneoplastic autoimmune symptoms may develop on rare occasions.
The genetic alterations present in TETs have been more deeply elucidated within the last 5 years. Similar to other tumor entities this has been feasible largely through the advent of next generation sequencing technologies. The results of these studies show a different pattern of molecular aberrations in thymomas and TCs, with only a few significantly and recurrently mutated genes. The perhaps most striking finding in thymomas is a very frequent point mutation, p.(Leu404His), in the general transcription factor IIi (GTF2I) gene, that so far has not been detected in other tumor entities (6). A further genetic hallmark is the mutation of epigenetic regulatory genes in TCs. The association of thymomas with MG has been linked to an increased gene copy number variation, and gene expression profiling identified overexpressed autoantigens (7). Unfortunately, the molecular profiling efforts have to date added only a few new therapeutic targets for currently available and approved drugs. Nevertheless, the novel insights into the genetic drivers of TETs form a strong basis for further studies on the biology and treatment of these tumors.
DNA alterations in TETs
GTF2I is a master genetic regulator in thymomas
The most frequently mutated gene in thymomas is GTF2I. It is mutated in 76–83% of types A and AB thymomas (6-8), less often in types B1, B2 and B3 subtypes and only in 8% of TCs (6,8). TETs with a GTF2I mutation exhibit a less aggressive clinical course than TETs with wild-type GTF2I, with 10 year-survival-rates of 96% and 70%, respectively. This is most likely caused by the higher GTF2I mutation prevalence in the indolent types A and AB thymomas (6).
The GTF2I gene recurrently carries the same point mutation on chromosome 7 at position 74146970T>A in TETs. The mutation leads to an exchange of the amino acid leucine by histidine on the protein level (p.Leu404His). The GTF2I gene codes for the transcription factor TFII-I, which regulates the expression of genes that influence the cell cycle (Figure 1) (9). The activation of TFII-I protein can be triggered by extracellular signals, such as the stimulation of B and T cell receptors and growth factor receptors. In mice the GTF2I gene is indispensable, because a knock-out of the gene causes death at an early embryonic stage (10).
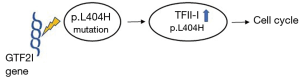
The p.(Leu404His) mutation changes an amino acid in a sequence segment that may function as a protein degradation mark (6). The mutation may thus increase the half-life of TFII-I and thereby support its intracellular accumulation (6,9). This assumption is fostered by the observation that GTF2I mutated TETs exhibit an increase of TFII-I protein in comparison with unmutated TETs, but without concomitant augmentation of mRNA expression (6). The transfection of a p.(Leu404His) mutated GTF2I gene into mouse NIH-3T3 cells increased their proliferation. However, in a soft-agar tumorigenicity assay, no difference between normal or mutated GTF2I transduced cells in their colony forming ability was observed (6). Therefore, mutant GTFI2 facilitates cell proliferation but does not exert a strong oncogenic effect.
The GTF2I p.(Leu404His) mutation is characteristic for TETs. For instance, it has not been observed in approximately 10,000 tumors of other entities that have been sequenced by the TCGA (7). However, 6% of angioimmunoblastic T cell lymphomas and less than one percent of other non-thymic tumor entities bear GTF2I missense mutations at other locations (11). Apart from that, a GTF2I-NCOA2 and a GTF2I-RARA gene fusion have been detected in an angiofibroma and in an acute promyelocytic leukemia, respectively (12,13). The GTF2I gene is located on chromosome 7 and the deletion of a region that spans GTF2I is present in patients with Williams-Beuren syndrome and a duplication has been linked with the Somerville-van der Aa syndrome (14). Both diseases exhibit malformations, mental retardation and disturbed behavior. Genome wide DNA sequence polymorphism studies suggested that certain GTF2I sequence variants contribute to the pathogenesis of autoimmune diseases such as rheumatoid arthritis, Sjögren’s syndrome and systemic lupus erythematosus (15-17). However, in thymomas no association of the GTF2I mutation with autoimmune diseases has been observed.
The recognition of the GFT2I p.(Leu404His) mutation as the most prominent genetic alteration in TETs helps to understand their pathogenesis. However, at present the GTF2I mutation cannot be therapeutically targeted. Even so, in clinical practice the identification of a GTF2I mutation may occasionally help pathologists to derive a diagnosis in small biopsies that are difficult to assess.
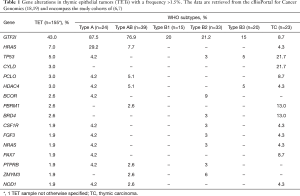
Irrespective of the GFT2I mutation, overall TETs have a low tumor mutational burden (TMB) with an average of 0.48 mutations per megabase. This is the lowest TMB among 21 other cancers that have been sequenced by The Cancer Genome Atlas (TCGA) (7). In addition to GTF2I only a few other genes were recurrently mutated in TETs at a frequency of at least 3% in a cohort of 155 reported cases (Table 1) (6,7,18,19). These genes were HRAS, TP53, CYLD, PCLO and HDAC4. HRAS mutations affected types A and AB thymomas in ten of eleven mutated TETs. Thus, HRAS is the second most frequently mutated gene in thymomas. TP53 and CYLD mutations do occur in thymomas but are more frequent in TCs (20,21). Mutations in the piccolo presynaptic cytomatrix gene PCLO and the chromatin modifier gene HDAC4 are observed in different thymoma subtypes and TCs with low frequencies and with no significant differences in incidence rates, although the number of published samples is too small to be able to draw firm conclusions.
Full table
TP53 and epigenetic modifier gene mutations are hallmarks of TCs
TCs exhibit a higher mutation rate than thymomas (20). The most frequently mutated gene is TP53. It has been altered in 26% of cases by mutation and frequently (32%) by hetero- or homozygous deletion as assessed by FISH (20,21). The presence of mutant TP53 has been associated with poorer overall survival in TC in some studies but this has not been confirmed by others (20-22).
A further genetic hallmark of TCs are recurrent mutations in chromatin modifying and epigenetic regulatory genes, particularly ASXL1, BAP1, DNMT3, SETD2, SMARCA4, TET2 and WT1 (20). ASXL1 is a polycomb chromatin binding protein, the tumor suppressor BAP1 encodes a histone deubiquitinase, DNMT3A represents a cytosine methyl transferase and SETD2 functions as a H3K36 trimethyl transferase. Furthermore, the SMARCA4 protein represents an ATPase of the SWI-SNF complex, TET2 functions as a methylcytosine dioxygenase and WT1 represents a positive regulator of DNMT3A and SMARCA4. The survival of TC patients with and without epigenetic regulatory gene mutations is similar (20). Another recurrently mutated gene in approximately 11% of TC is CYLD deubiquitinase, which functions as a negative regulator of the NFKB pathway (6,20).
The tyrosine kinase KIT is expressed by 80–86% of thymic squamous carcinomas, but infrequently by non-TCs (23,24). It therefore represents a useful immunohistochemical marker in the histopathologic diagnosis of TC (25). KIT mutations, however, affect only 6–9% of TC, and only in these mutated cases KIT constitutes a potential target for treatment with tyrosine kinase inhibitors (TKIs) (20,21).
Gene copy number changes in TETs
Gene copy number aberrations (CNAs) are rare in A and AB thymomas but enriched in B2 and B3 thymomas and TCs (6,7,26). If present, most are large-scale, chromosome arm level CNAs (6,7,26). Deletions involve most frequently the chromosome arms 3p, 6p, 6q, 13q, 16q and 17p (6,7,26,27). Gains of chromosomal material are most common for 1q, 17q and 18 (7,26). In TC the loss of chromosome 16q is most prominent and reported for 67% of cases (7,26). Although no single gene on 16q that is solely crucial for TC pathogenesis has been pinpointed, several candidate tumor suppressor genes are mapped to this chromosome arm, namely CBFB, CDH1, CDH11, CTCF, CYLD and ZFHX3.
A further frequently mutated and/or deleted gene in TC is the tumor suppressor CDKN2A located at 9p21.3. CDKN2A mutations, mostly truncating, have been reported for 11% of TC and a hetero- or homozygous deletion has been observed in 38% of cases in a cohort of 35 TC (21,27). The CDKN2A gene codes for p16INK4A and p14ARF. The two proteins are generated by alternative RNA splicing. The p16INK4A inhibits mitosis by antagonizing cyclin dependent kinases 4 and 6. On the other hand, p14ARF activates the anti-oncogene TP53. Therefore, a CDKN2A deletion or inactivation by mutation may result in a stimulation of cyclin dependent kinases with subsequent promotion of cell proliferation.
Genetic aberrations in neuroendocrine thymic tumors
The WHO classification of thymic NETs is similar to pulmonary NETs and distinguishes carcinoid (typical and atypical), large cell neuroendocrine carcinoma and small cell carcinoma (1,28). So far little is known about the genetic aberrations in thymic NETs, which is likely due to the rarity of these tumors. However, some 25% of thymic carcinoids are associated with multiple endocrine neoplasia-1 (MEN1), a hereditary tumor syndrome that is caused by mutation of the MEN1 gene. In contrast, only 2–8% of patients with MEN1 develop a thymic carcinoid.
A recent study analyzed 63 thymic NETs by low coverage whole genome sequencing to determine gene copy number instabilities (CNI) (29). The report differentiated three groups of thymic NETs with low, intermediate and high CNI. The CNI low and intermediate tumors encompassed in addition to carcinoids also some large cell neuroendocrine carcinomas. On the other hand, the group with high CNI reflected all small cell carcinomas and most large cell neuroendocrine carcinomas but contained also a few atypical carcinoids. Therefore, histopathology failed to faithfully predict the CNI grouping. The authors of the study have thus concluded that thymic NETs separate into three genetic clusters which are not satisfactorily recognized by the WHO classification. A morphomolecular grading system for thymic NETs was proposed instead of a mere histologic classification for patient stratification and prognostication (29). However, such a grading system would be difficult to implement in clinical practice, because of the requirement of low coverage whole genome sequencing and assay standardization among diagnostic laboratories.
The transcriptome of TETs
RNA expression profiles mirror the histopathologic classification of TETs
RNA sequencing (RNA-seq) of 12 thymomas and one TC revealed gene transcription profiles that correlated well with the WHO histopathologic subtyping (30). A similar RNA-seq analysis of a larger cohort of 117 TETs within the TCGA project that integrated RNA-seq, gene copy number variation, DNA methylation, and protein array findings revealed also a strong correlation of gene expression profiles with histologic subtyping (7). The study differentiated four molecular subgroups. Group 1 were mainly type B thymomas, group 2 TCs, group 3 primarily type AB thymomas and group 4 constituted types A and AB thymomas. RNA-seq detected only a few gene fusions in TETs and none of them was recurrent or involved the GTF2I gene (6,7). Lee et al. evaluated in silico the TET data of TCGA and suggested also the separation of TETs into four groups, although with different molecular features defined by GTF2I mutation, a normal GTF2I phenotype with expression of T cell signaling genes, and TETs with chromosomal stability or chromosomal instability (31). Obviously, the genetic TET grouping by Lee et al. cannot be predicted by histology, reminiscent to molecular subtyping efforts in other tumors, e.g. bladder cancer (31,32).
The C19MC miRNA cluster characterizes A and AB thymomas
The C19MC miRNA cluster is strongly expressed in types A and AB thymomas, but virtually absent in type B thymomas and TCs (21,30). It is a large imprinted miRNA cluster on chromosome 19q13.42 that is normally expressed only during embryonic development and is present in the placenta but silenced in adult tissues. An exception are medullary and cortical thymic epithelial cells which do express C19MC (33). Therefore, the expression of C19MC in A and AB thymomas may reflect a physiological state, and expression is likely silenced in type B thymomas and TCs by a mechanism that is currently not known, but might involve promoter methylation (34). In types A and AB thymomas the miRNAs of the C19MC cluster may activate the PI3K/AKT pathway as proposed by Radovich et al. (30).
The C19MC miRNA cluster is expressed also in a few non-thymic malignancies. In the rare and aggressive brain tumors ETANTRs (embryonal tumors with abundant neuropil and true rosettes) and ETMRs (embryonal tumors with multilayered rosettes) the C19MC cluster is amplified or fused with the TTYH1 gene promoter, which results in overexpression (35,36). Additionally, the C19MC cluster is expressed in adenomas of the thyroid and parathyroid, tamoxifen non-responsive breast tumors and liver cell carcinomas and mesenchymal hamartomas. The mechanism of overexpression in these tumors are variable and encompass gene amplification, chromosomal translocation, and hypomethylation (37-40).
The expression of a further miRNA cluster, called C14MC and located on chromosome 14q32, is also reduced in TCs, albeit not all miRNAs of the cluster are silenced (21). The C14MC cluster has been regarded to possess a tumor suppressive function in GIST and glioma (41,42). The downregulation of anti-oncogenic C14MC miRNAs might thus similarly promote the genesis of TCs.
The reports on non-clustered miRNA expression in TETs are more heterogenous and therefore difficult to integrate into a common pattern (21,43,44). This may be caused by the use of different techniques (microarrays, next generation sequencing/NGS) and varied TET subtype composition and size of the study cohorts. Enkner et al. observed in association with NGS the upregulation of mainly oncogenic miR-21, miR-9-3 and miR-375 and the reduction of putative tumor suppressive miR-34b, miR-34c, miR-130a and miR-195 in TCs as compared to type A thymomas (21). Notably, in non-small cell lung cancer TP53 downregulates PD-L1 via miR-34, and functional loss of TP53 and consecutive silencing of miR-34 increases PD-L1 and thereby supports immune evasion (45). TCs express PD-L1 more frequently than type A thymomas, and this might likewise be caused by a decrease of miR-34.
Ganci et al. compared the miRNA expression of 54 TETs with normal thymic tissue by microarray hybridization and identified 87 differently expressed miRNAs (43). Similar to the observation by Enkner et al. an upregulation of oncogenic miR-21-5p was recognized among the most significantly deregulated miRNAs. In addition, the silencing of the tumor suppressive miR-145-5p was found to be significant, and an epigenetic regulation of miR-145-5p expression, previously demonstrated in other tumor entities, was revealed in TETs as well (43,44).
The molecular pathogenesis of MG
About 30% of thymoma patients exhibit an autoimmune disorder, in particular MG. The autoantibodies are directed against muscle specific antigens, mainly the acetylcholine receptor (AChR) in the postsynaptic muscle membrane (4,46). Localized or general muscle weakness is the predominant symptom. Although the disease-inducing antibodies have been well characterized, the pathogenetic mechanisms triggering the autoimmune disease are less understood. However, similar to other autoimmune diseases an association of MG with certain major histocompatibility complex (MHC)—classes has been observed (4).
The recent genomic characterization of thymomas by the TCGA have revealed that thymomas with MG exhibit a higher prevalence of gene copy aberrations than thymomas in the absence of MG (7). However, which of the deletions and gains of chromosomal material are crucial for the development of MG could not be clarified, because none of the gene CNAs was significantly accumulated in the thymomas with MG. Furthermore, MG did not segregate with any gene mutation and DNA methylation or gene expression pattern (7). Surprisingly, thymomas associated with MG do not express complete prototypical target molecules of autoantibodies such as AChR, titin, and muscle ryanodine proteins 1 and 2. Expression of the AChR alpha subunit was only 3.0-fold higher in 32 thymomas with MG than in 72 tumors without MG. By contrast, expression of NEFM (neurofilament medium) which shares sequence similarities with AChR, titin, and ryanodine receptors was 23.8-fold higher in MG positive compared with MG negative thymomas (7). Furthermore, the neuronal ryanodine receptor 3, which shares homology with muscle ryanodine receptors 1 and 2, was 5.5-fold upregulated in MG positive thymomas. In summary, MG in thymoma concurs with an increased intratumoral expression of genes that share sequence homologies with autoantibody targets.
Targeted therapies in TETs
Surgical resection is the primary therapy for TETs. In addition, for unresectable or incompletely resected tumors, chemo- and radiotherapy are used (47). At present no targeted therapy for TETs is firmly established. This is brought about by the low number of recurrent and actionable mutations and the rarity of TETs, which makes clinical trials difficult. Nevertheless, activating mutations in the KIT oncogene, which are present in approximately 5% of TCs, have been explored as a therapeutic target for TKIs with encouraging results in a few patients (48,49). However, more informative clinical studies with a larger number of KIT mutated patients are needed. Approximately 80% to 86% of TCs express KIT protein, but this does not correlate with their response to TKIs (25,47). Therefore, only TCs with an activating KIT mutation are potential candidates for a TKI therapy.
The genomic profiling of TETs revealed additional candidate molecular targets (6,7,20,21). Amongst them the miRNA cluster C19MC, which is a unique feature of types A and AB thymomas (21,30). The cluster activates the PI3K pathway which might be effectively blocked in the rare incidences of recurrent types A and AB thymomas. The common mutation of genes that modify the epigenetic signatures of DNA and chromatin in TCs could offer an additional chance for therapeutic intervention (20).
Immune checkpoint inhibitors may also evolve as a therapeutic option in clinically advanced TETs. PD-L1 is frequently expressed by tumor cells, particularly in B3 thymomas and TCs (21,50-53). In 40 patients with recurrent TCs who had progressed after at least one line of chemotherapy the immune-checkpoint inhibitor pembrolizumab achieved complete or partial response in 22.5% and stable disease in 53% of patients (54). Another phase II clinical trial with pembrolizumab enrolled 26 TC and seven thymoma patients (55). Five (19.2%) of the TCs exhibited a partial response and 14 (53.8%) had stable disease. Of the seven thymoma patients, two achieved partial response and five stable disease. In both studies high PD-L1 protein expression was associated with better response to pembrolizumab. Although the observed responses of TETs to immune checkpoint inhibitor therapy are promising, the high incidence rate of severe autoimmune adverse advents that has been observed is of major concern, in particular in the thymoma patients (54,55).
Conclusions
In summary, exemplary prominent insights derived from molecular profiling are the identification of GTF2I as an important gene for the development of TETs, the discovery of frequent epigenetic regulatory gene mutations in TCs and the recognition of the large miRNA cluster C19MC in types A and AB thymomas. The translation of the molecular insights into a clinical benefit for TET patients will need the development of novel targeted medicines that address different molecules and the organization of multi-center trials, because of the diversity and infrequence of recurrent genetic alterations and the rareness of TETs.
Acknowledgments
We thank Gertrude Krainz for proof-reading the manuscript.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Dragana Jovanovic and Semra Bilaceroglu) for the series “Thymoma” published in Journal of Thoracic Disease. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2019.12.52). The series “Thymoma”was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Burke AP, et al. editors. WHO classification of tumours of the lung, pleura, thymus and heart. IARC Press, Lyon; 2015.
- Engels EA. Epidemiology of Thymoma and Associated Malignancies. J Thorac Oncol 2010;5:S260-5. [Crossref] [PubMed]
- Fonseca AL, Ozgediz DE, Christison-Lagay ER, et al. Pediatric thymomas: report of two cases and comprehensive review of the literature. Pediatr Surg Int 2014;30:275-86. [Crossref] [PubMed]
- Gilhus NE. Myasthenia Gravis. N Engl J Med 2016;375:2570-81. [Crossref] [PubMed]
- Marx A, Ströbel P, Weis CA. The pathology of the thymus in myasthenia gravis. Mediastinum 2018;2:66. [Crossref]
- Petrini I, Meltzer PS, Kim IK, et al. A specific missense mutation in GTF2I occurs at high frequency in thymic epithelial tumors. Nat Genet 2014;46:844-9. [Crossref] [PubMed]
- Radovich M, Pickering CR, Felau I, et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell 2018;33:244-58.e10. [Crossref] [PubMed]
- Feng Y, Lei Y, Wu X, et al. GTF2I mutation frequently occurs in more indolent thymic epithelial tumors and predicts better prognosis. Lung Cancer 2017;110:48-52. [Crossref] [PubMed]
- Roy AL. Pathophysiology of TFII-I: Old Guard Wearing New Hats. Trends Mol Med 2017;23:501-11. [Crossref] [PubMed]
- Enkhmandakh B, Makeyev AV, Erdenechimeg L, et al. Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. Proc Natl Acad Sci U S A 2009;106:181-6. [Crossref] [PubMed]
- Vallois D, Dobay MP, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 2016;128:1490-502. [Crossref] [PubMed]
- Arbajian E, Magnusson L, Mertens F, et al. A novel GTF2I/NCOA2 fusion gene emphasizes the role of NCOA2 in soft tissue angiofibroma development. Genes Chromosomes Cancer 2013;52:330-1. [Crossref] [PubMed]
- Li J, Zhong HY, Zhang Y, et al. GTF2I-RARA is a novel fusion transcript in a t(7;17) variant of acute promyelocytic leukaemia with clinical resistance to retinoic acid. Br J Haematol 2015;168:904-8. [Crossref] [PubMed]
- Van der Aa N, Rooms L, Vandeweyer G, et al. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet 2009;52:94-100. [Crossref] [PubMed]
- Li Y, Zhang K, Chen H, et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjogren's syndrome at 7q11.23. Nat Genet 2013;45:1361-5. [Crossref] [PubMed]
- Sun C, Molineros JE, Looger LL, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet 2016;48:323-30. [Crossref] [PubMed]
- Kim K, Bang SY, Ikari K, et al. Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis. Sci Rep 2016;6:27563. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Wang Y, Thomas A, Lau C, et al. Mutations of epigenetic regulatory genes are common in thymic carcinomas. Sci Rep 2014;4:7336. [Crossref] [PubMed]
- Enkner F, Pichlhöfer B, Zaharie AT, et al. Molecular Profiling of Thymoma and Thymic Carcinoma: Genetic Differences and Potential Novel Therapeutic Targets. Pathol Oncol Res 2017;23:551-64. [Crossref] [PubMed]
- Moreira AL, Won HH, McMillan R, et al. Massively parallel sequencing identifies recurrent mutations in TP53 in thymic carcinoma associated with poor prognosis. J Thorac Oncol 2015;10:373-80. [Crossref] [PubMed]
- Pan CC, Chen PC, Chiang H. KIT (CD117) is frequently overexpressed in thymic carcinomas but is absent in thymomas. J Pathol 2004;202:375-81. [Crossref] [PubMed]
- Nakagawa K, Matsuno Y, Kunitoh H, et al. Immunohistochemical KIT (CD117) expression in thymic epithelial tumors. Chest 2005;128:140-4. [Crossref] [PubMed]
- Moser B, Schiefer AI, Janik S, et al. Adenocarcinoma of the thymus, enteric type: report of 2 cases, and proposal for a novel subtype of thymic carcinoma. Am J Surg Pathol 2015;39:541-8. [Crossref] [PubMed]
- Zettl A, Strobel P, Wagner K, et al. Recurrent genetic aberrations in thymoma and thymic carcinoma. Am J Pathol 2000;157:257-66. [Crossref] [PubMed]
- Petrini I, Meltzer PS, Zucali PA, et al. Copy number aberrations of BCL2 and CDKN2A/B identified by array-CGH in thymic epithelial tumors. Cell Death Dis 2012;3:e351. [Crossref] [PubMed]
- Bohnenberger H, Dinter H, König A, et al. Neuroendocrine tumors of the thymus and mediastinum. J Thorac Dis 2017;9:S1448-57. [Crossref] [PubMed]
- Dinter H, Bohnenberger H, Beck J, et al. Molecular Classification of Neuroendocrine Tumors of the Thymus. J Thorac Oncol 2019;14:1472-83. [Crossref] [PubMed]
- Radovich M, Solzak JP, Hancock BA, et al. A large microRNA cluster on chromosome 19 is a transcriptional hallmark of WHO type A and AB thymomas. Br J Cancer 2016;114:477-84. [Crossref] [PubMed]
- Lee HS, Jang HJ, Shah R, et al. Genomic Analysis of Thymic Epithelial Tumors Identifies Novel Subtypes Associated with Distinct Clinical Features. Clin Cancer Res 2017;23:4855-64. [Crossref] [PubMed]
- Bertz S, Eckstein M, Stoehr R, et al. Urothelial Bladder Cancer: An Update on Molecular Pathology with Clinical Implications. Eur Urol Suppl 2017;16:272-94. [Crossref]
- Ucar O, Tykocinski LO, Dooley J, et al. An evolutionarily conserved mutual interdependence between Aire and microRNAs in promiscuous gene expression. Eur J Immunol 2013;43:1769-78. [Crossref] [PubMed]
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 2010;19:3566-82. [Crossref] [PubMed]
- Korshunov A, Remke M, Gessi M, et al. Focal genomic amplification at 19q13.42 comprises a powerful diagnostic marker for embryonal tumors with ependymoblastic rosettes. Acta Neuropathol 2010;120:253-60. [Crossref] [PubMed]
- Kleinman CL, Gerges N, Papillon-Cavanagh S, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet 2014;46:39-44. [Crossref] [PubMed]
- Rippe V, Dittberner L, Lorenz VN, et al. The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS One 2010;5:e9485. [Crossref] [PubMed]
- Vaira V, Elli F, Forno I, et al. The microRNA cluster C19MC is deregulated in parathyroid tumours. J Mol Endocrinol 2012;49:115-24. [Crossref] [PubMed]
- Flor I, Bullerdiek J. The dark side of a success story: microRNAs of the C19MC cluster in human tumours. J Pathol 2012;227:270-4. [Crossref] [PubMed]
- Kapur RP, Berry JE, Tsuchiya KD, et al. Activation of the chromosome 19q microRNA cluster in sporadic and androgenetic-biparental mosaicism-associated hepatic mesenchymal hamartoma. Pediatr Dev Pathol 2014;17:75-84. [Crossref] [PubMed]
- Haller F, von Heydebreck A, Zhang JD, et al. Localization- and mutation-dependent microRNA (miRNA) expression signatures in gastrointestinal stromal tumours (GISTs), with a cluster of co-expressed miRNAs located at 14q32.31. J Pathol 2010;220:71-86. [Crossref] [PubMed]
- Lavon I, Zrihan D, Granit A, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol 2010;12:422-33. [Crossref] [PubMed]
- Ganci F, Vico C, Korita E, et al. MicroRNA expression profiling of thymic epithelial tumors. Lung Cancer 2014;85:197-204. [Crossref] [PubMed]
- Bellissimo T, Ganci F, Gallo E, et al. Thymic Epithelial Tumors phenotype relies on miR-145-5p epigenetic regulation. Mol Cancer 2017;16:88. [Crossref] [PubMed]
- Cortez MA, Ivan C, Valdecanas D, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2015. [Crossref] [PubMed]
- Gilhus NE, Skeie GO, Romi F, et al. Myasthenia gravis - autoantibody characteristics and their implications for therapy. Nat Rev Neurol 2016;12:259-68. [Crossref] [PubMed]
- Merveilleux du Vignaux C, Maury JM, Girard N. Novel Agents in the Treatment of Thymic Malignancies. Curr Treat Options Oncol 2017;18:52. [Crossref] [PubMed]
- Strobel P, Hartmann M, Jakob A, et al. Thymic carcinoma with overexpression of mutated KIT and the response to imatinib. N Engl J Med 2004;350:2625-6. [Crossref] [PubMed]
- Bisagni G, Rossi G, Cavazza A, et al. Long lasting response to the multikinase inhibitor bay 43-9006 (Sorafenib) in a heavily pretreated metastatic thymic carcinoma. J Thorac Oncol 2009;4:773-5. [Crossref] [PubMed]
- Girard N. Use of immune checkpoint inhibitors in thymic epithelial tumors. Mediastinum 2018;2:34. [Crossref]
- Sekine I, Aida Y, Suzuki H. Expression patterns and prognostic value of programmed death ligand-1 and programmed death 1 in thymoma and thymic carcinoma. Mediastinum 2018;2:54. [Crossref]
- Sakane T, Murase T, Okuda K, et al. A comparative study of PD-L1 immunohistochemical assays with four reliable antibodies in thymic carcinoma. Oncotarget 2018;9:6993-7009. [Crossref] [PubMed]
- Owen D, Chu B, Lehman AM, et al. Expression Patterns, Prognostic Value, and Intratumoral Heterogeneity of PD-L1 and PD-1 in Thymoma and Thymic Carcinoma. J Thorac Oncol 2018;13:1204-12. [Crossref] [PubMed]
- Giaccone G, Kim C, Thompson J, et al. Pembrolizumab in patients with thymic carcinoma: a single-arm, single-centre, phase 2 study. Lancet Oncol 2018;19:347-55. [Crossref] [PubMed]
- Cho J, Kim HS, Ku BM, et al. Pembrolizumab for Patients With Refractory or Relapsed Thymic Epithelial Tumor: An Open-Label Phase II Trial. J Clin Oncol 2019;37:2162-70. [Crossref] [PubMed]


