
Identification of somatic copy number variations in plasma cell free DNA correlating with intrinsic resistances to EGFR targeted therapy in T790M negative non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer death worldwide. In the past decades, significant improvements have been made for lung cancer treatment, based on discoveries of molecular biomarkers and disease progression mechanisms. A landmark of these improvements is epidermal growth factor receptor (EGFR) targeting therapy for patients with advanced lung cancer bearing EGFR activating mutations (1,2). However, clinical outcomes for lung cancer still remains unsatisfactory, with 5-year survival rate of less than 20% (3).
Clinical studies have revealed that lung cancer with activating EGFR mutations that are initially responsive to EGFR-tyrosine kinase inhibitors (EGFR-TKIs) will develop acquired resistance to TKI after a median progression-free survival (PFS) of 10–16 months (2,4). On the other hand, 20–30% of patients with non-small cell lung cancer (NSCLC) harboring activating EGFR mutation show no objective tumor regression to initial EGFR-TKI treatment, and this subgroup of NSCLC has been defined as with intrinsic or primary resistance to EGFR-TKIs.
Studies have revealed that the secondary T790M mutation of EGFR is the major cause of the acquired EGFR-TKI resistance in lung cancer (5). The third-generation of EGFR-TKI, aiming targeting the acquired EGFR T790M resistance mutation, has thus been developed. Clinical studies showed that osimertinib, a first third-generation of EGFR-TKI received FDA and EMA approval, demonstrated significant clinical effects with 70% objective response rate and 10 months progression free survival in metastatic EGFR-mutant NSCLC patients failure to first-generation EGFR-TKIs (6). Other causes for the acquired EGFR-TKI resistance in lung cancer include c-MET amplification (7), HER2 and PIK3CA mutation (8).
Very little is known about intrinsic resistance of EGFR-TKI, however, especially for the lung cancers that are with activating EGFR mutation but negative for T790M. Of note, most studies focused on single gene alteration, but it is needed to be indicated that multiple resistance mechanisms may co-exist because of tumor heterogeneity (9,10). Therefore, novel perspective study for EGFR-TKI resistance is needed.
Genomic instability involves a transient phase of tetraploidization. Tetraploid cells can undergo asymmetric cell division or chromosome loss, leading to increased tumor heterogeneity and multidrug resistance (11). Experimental evidence further revealed that chromosomal instability enables tumor adaptation with aneuploid fitness landscape (12,13). In lung cancer cells, chromosome 7 aneuploidy was found to be one of the most important events for cancer development. These deregulations or variations of Chromosome 7 can be frequently detected in malignant lung cancer and pre-cancerous lesions cells, or even in lung bronchial cells, but not in health lung tissues cells (14-16). Chromosome aneuploidy detection has been used for prenatal tests through plasma cell free DNA, with minimal false positives and false negatives (17). Similar to fetal tissues and cells, tumors also keep shedding DNA into peripheral blood stream. As such, technology of ctDNA (circulated tumor DNA) has been successfully applied in clinic to detect biomarkers or cancer somatic mutations such as EGFR mutations for predicting potential benefits of targeted therapies. In addition, detection of chromosomal copy number changes with ctDNA technology has also been reported in patients with breast cancer (18), hepatocellular carcinoma (19) and lung cancer (20).
The criteria for clinical defining intrinsic EGFR-TKI resistance for lung cancer have not been established. Jackman et al. (21) proposed criteria for acquired resistance of EGFR-TKI in lung cancer patients with mutant EGFR, including that patients achieve a partial or complete response or develop a stable disease in response to EGFR-TKI monotherapy (>6 months). Of note, patients with PFS less than 6 months were recruited in the study, and thus the lung cancer of these patients potentially harbored intrinsic resistant to EGFR-TKI treatment.
Low pass whole genome sequencing approach with an optimized bioinformatics pipeline, ultra-sensitive chromosomal aneuploidy detector (UCAD), were used to screen chromosomal aneuploidy, especially chr7 aneuploidy, by using plasma cell free DNA in EGFR targeted therapy intrinsic resistant patients.
Methods
Patients
Thirty-one lung cancer patients and ten health volunteers were enrolled in this study. All lung cancer patients relapsed after EGFR-TKI treatment. The protocol of the study was approved by the Institutional Review Board of Hangzhou First People’s Hospital (No. HZFH CA15-02). All recruited patients and health volunteers have signed a written informed consent.
Sample collection and DNA extraction
Blood samples were collected within 14 days after the development of TKI resistance as assessed by the physician according to the Jackman criteria (21) and before the start of the following treatment. Approximately 10–15 mL of peripheral blood was collected in a cell-free DNA protection vacuum tube (AmoyDx, Xiamen, Fujian, China), which contains a cell-free DNA protection reagent to keep DNA stable for 7 days at 4–25 °C. Blood samples were transported to the Center for Translational Medicine of Hangzhou First People’s Hospital within 36 hours for further processing. For DNA extraction, the blood samples were centrifuged at 2,500 ×g for 10 minutes at 4 °C, and the supernatant was transferred to a new tube for further centrifugation at 15,800 ×g for 15 minutes at 4 °C. The collected plasma supernatant was then stored at −80 °C. Cell-free DNA from 1.5 mL plasma was extracted with a QIAamp Circulating Nucleic Acid kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany).
ARMS assays for testing EGFR T790M mutations
The EGFR T790M mutational status was determined by ARMS (amplification refractory mutation system) with the ADx-ARMS kit (AmoyDx, Xiamen, China). EGFR mutations in plasma ctDNA were detected by using the plasma EGFR detection kit on qPCR platform. All experiments and genotype calling were performed according to the manufacturer’s instructions (22).
Next generation sequencing
Next generation sequencing was performed as previously described (19,23). Briefly, DNA was fragmented into an average size of 300 bp (cfDNA without fragmentation), and 100 ng of fragmented genomic DNA (or 10 ng for cfDNA) was used for preparation of sequencing libraries (NEBnext Ultra II). Eight bp barcoded sequencing adaptors were then ligated to the DNA fragments and the DNA templates were amplified by PCR. Purified sequencing libraries were massively parallel sequenced by Illumina HiSeq Xten platform. 4G sequencing raw data per sample were filtered and aligned to the human reference genome.
Gene-level copy number analyses
Chromosome copy number aberrations (CNAs) were determined with the Ultrasensitive Chromosomal Aneuploidy Detector (UCAD) pipeline. Sequencing coverage for each 200 K bin was calculated followed by GC normalization. The sequencing coverage were further normalized by controls samples. The Z-score for each bin was calculated by formula,
|
| [1] |
where Ctest and Ccontrol are the coverage of the bin. The normalized bin values were sent to segmentation calls by algorithm circular segmentation algorithm as provided by R package DNAcopy. Samples with standard deviation of copy ratios between the adjacent bins >30 for genome-wide results were considered as with poor-quality sequence data, and these samples were excluded from this study.
Statistical analysis
R package ‘DNACopy’ was used for analysis of copy number changes. A P value of <0.05 was considered as statistically significant binary segmentation. Absolute segment value is used for further analysis. The sensitivity and specificity of UCAD were estimated by ROC curves. The chi-square test was used for categorical variables. OS was calculated from the time of development of TKI resistance to the time of death of any reason or last follow-up. Survival curves were constructed using the Kaplan-Meier method and compared using the log-rank test. All statistical analyses were performed using SPSS 17.0.
Results
Patient characterization
In this study, thirty-one enrolled lung cancer patients were collected from a previous study (clinical trial NCT02418234) which aimed to analyze the association of clinical mode and plasma T790M mutation in the relapsed patients after treatment with first generation TKI (24). Of them, 18 patients were with PFS less than 6 months and thus the tumors were considered to be with primary resistance to EGFR-TKI, 13 patients were with PFS longer than 36 months and were defined with acquired EGFR-TKI resistances. As shown in Table 1, plasma T790M mutation is negatively detected in all patients by using Amplification Refractory Mutation System PCR. No statistical differences were observed for gender and EGFR baseline mutations between the two groups with primary and acquired EGFR-TKI resistance. Of note, clinical trial NCT02418234 involved 307 patients, and only these thirty-one plasma T790M negative patients shown here were found the PFS less than 6 months or more than 36 months, and our data showed that patients with primary resistance tend to be 10-year younger than that with acquired resistance. With 60-year as a cutoff value, we did not find statistical significance for the incidence of patients between these two groups.
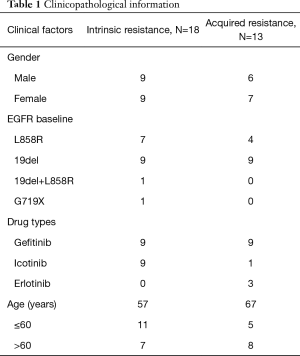
Full table
Cell-free DNA whole genome copy number profiling
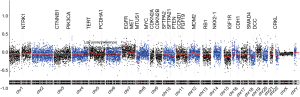
In this study, all raw sequencing reads were mapped to human reference genome hg19, and genomic coverage was counted by using software samtools mpileup. With these setting, we counted the average coverage for each 200k bin, and determined the significant genomic breakpoints with using circular binary segmentation algorithm. Our results showed that in all 31-tumor specimen, when compared to normal control, chromosomal breakpoints were commonly detected on centromere regions and chromosomal arm coverage imbalances were found on chromosome 1, 7 and 8 by visual inspections. In addition, chromosome 1 short arm (1p) was found with coverage lower than normalized average (as calculated as 0), indicating 1p copy loss, and chromosome 1 long arm (1q) was found with coverage higher than the normalized average, indicating 1q copy number gains. Similarly, chromosome 7 short-arm was found with higher coverage comparing to long arm, indicating chromosomal arm copy gains. Analysis further revealed a statistically significant focal amplification on chromosome 7p11.2, a loci where EGFR located. We also noted a copy number gain peak around 7q31.2. Of interest, oncogene MET, a well-known cancer driver, is located in this loci (Figure 1). In these analyses, the statistical significance of copy gain/loss was calculated by Student t-test by comparing chr1p 200-bin coverage values against that of health controls.
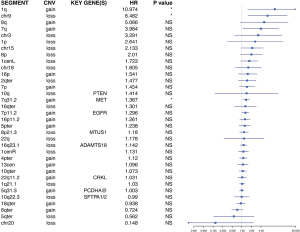
Comprehensive analysis further demonstrated statistically significant chromosome arm level changes between tumors with intrinsic resistance and with acquired resistance. Figure 2 shows a heatmap illustrating the most significant genomic changes detected. These include genetic events of 7p gains (Z ≥3 in 8/18, 44.4%), 1q heavy gains (Z ≥6 in 5/18, 27.8%), 7q31.2 gains (Z ≥3 in 2/18, 11.1%) and 7p11.2 heavy gains (Z ≥6 in 5/18, 27.8%), frequent chromosome losses of chr18 (7/18, 38.9%), chr9 (6/18, 33.3%), chr3 (6/18, 33.3%), 16p (1/18, 5.56%), 1p (7/18, 38.9%), chr20 (10/18, 55.6%), 22q (4/18, 22.2%), 10q (8/18, 44.4%), 8p (3/18, 16.7%) and chr15 (6/18, 33.3%) and chromosomal gains of 7q (5/18, 27.8%), 7p (8/18, 44.4%), 8q (4/18, 22.2%) and 1q (6/18, 33.3%) that are exclusively found in the group with intrinsic resistance of EGFR-TKI. On the other hand, genetic events with copy loss in chr18 (4/13, 30.8%), chr9 (1/13, 7.69%), chr3 (4/13, 30.8%), 16p (2/13, 15.4%), 1p (3/13, 23.1%), chr20 (6/13, 46.1%), 22q (1/13, 7.69%), 10q (6/13, 46.1%), 8p (1/13, 7.69%) and chr15 (2/13, 15.4%), and genetic events with copy gain in 7q (3/13, 23.1%), 8q (1/13, 7.69%) and 1q (1/13, 7.69%) were detected in the specimen from patient with lung cancer that are T790M negative but with acquired resistance to EGFR-TKI.
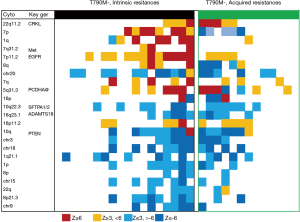
In addition, non-arm level copy number gains in 7p11.2, 22q11.2, 16p11.2 and 5q31.3, including focal amplifications, were detected in specimen from patients with lung cancer that were defined as with intrinsic resistance of EGFR-TKI with frequencies of 66.7% (12/18), 22.2% (4/18), 33.3% (6/18) and 33.3% (6/18), and in patients with lung cancer that were with acquired resistance with frequencies of 30.8% (4/13), 38.5% (5/13), 53.8% (7/13) and 38.5% (5/13), respectively (Figure 2). Of interest, all these amplified locations are linked with potential lung cancer oncogenes. For example, lung cancer oncogene EGFR locates in 7p11.2 (1); CRKL, a recently identified lung cancer driver predicting the relapse of patient after TKI treatment (25), locates in 22q11.2; ADAMTS18 which contributes to lung cancer development (26) locates in 16p11.2. PCDHA, methylation of this gene has been reported to be involved in multiple caner development (27), locates in 5q31.3.
chr7p gains is a frequent event detected in patients with lung cancer that are EGFR T790M negative but with intrinsic TKI resistance
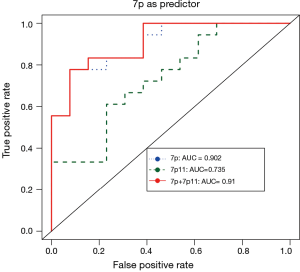
We next examined the potential correlations of chromosomal copy number changes with TKI resistances. Of interest, our data showed that chromosome 7 copy number gain significantly correlates with intrinsic TKI resistances: 7p11.2 (EGFR loci) gains was detected in 66.7% (12/18) of lung cancer patients with intrinsic TKI resistance while it is found in only 30.7% (4/13) of patients with acquired TKI resistance; 7p arm gains were found in 44.4% (8/18) of patients with intrinsic resistance, and no such event was detected in patients with acquired TKI resistance (Fisher exact test, P=0.009). However, no statistical significance was found for the other frequent detected genomic events as listed in Table 2, including 8q gain, 8p21.3 loss and 1q gain. The operating characteristic (ROC) analysis further revealed that chr7p copy number gains of chr7p can serve as a predictor for distinguishing intrinsic resistance from acquired resistance for lung cancer in response to EGFR-TKI (Figure 3).
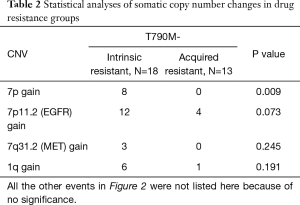
Full table
All the other events in Figure 2 were not listed here because of no significance.
EGFR copy number as a predictor for EGFR-TKI intrinsic resistance in T790M negative patients. AUC curves for 7p gains to predict intrinsic resistance. The performance of 7p11 is indicated in green dash line. The performance of 7p is indicated in blue dot line. The performance of combining 7p and 7p11 is indicated in red line. The combining product was estimated by 10.7×7p+0.619×7p11. The coefficients were estimated by logistical regression Category~logit (7p+7p11), where Category is the category label of a sample, which is either ‘intrinsic resistance’ or ‘acquired resistance’.
Copy number changes correlate with overall survival (OS) after TKI resistances
We also determined the potential correlations between these observed chromosome changes and OS of patient (Figure 4). For this, patients were grouped with short OS (≤6 months), medium OS (6–12 months) and long OS (≥12 months). We found that chr9 loss and 1q gain significantly correlated with shorter OS (trend tests, P=0.020 and 0.029 respectively). In particular, patients with 1q gains detected had shorter OS (medium average of OS for 3.4 months) when compared to patients with 1q silent of lung cancer (medium average of OS for 22.2 months, hazard ratio =10.97, log rank test P=0.029). The similar association of chromosomal changes with poor OS were also observed with chr9 loss (hazard ratio =6.48, P=0.020), and 7q31.2 gain (hazard ratio =1.37, P=0.031) where oncogenes CDKN2A/B and MET locate (28,29). No such correlations were found for other genetic events such as 8p loss, 1p loss, 8p21.3 loss, 7q gain, 8q gain and 16p11.2 loss, however.
Discussion
EGFR-targeted therapy has shown superior efficacy for patients with EGFR mutation. However, about 20% to 30% patients with advanced stage lung cancers that have EGFR activating mutation show intrinsic resistance to TKI. Understanding the mechanism of the intrinsic EGFR-TKI resistance is thus important for helping improve clinical practice for lung cancer.
In this study, we identified multiple somatic copy number variations (CNV) of chromosome through plasma cfDNA sequencing. These variants include chromosome arm level changes, focal amplifications and deletions. In mammalian cells, chromosome mis-segregation can result in chromosomal arm level changes, which affects the gene structure of many tumor genes, such as PTEN deletions on chr10 and SMAD4 deletions on chr18 (30,31). Focal amplifications, or focal chromosomal CNAs, have been discovered in cancer as critical genetic events of cancer driver gene activation resulting from many selection events during the evolution of cancer genomes (32). Our results showed that chromosomal breakpoints on centromere regions and CNV of chromosomes on Chromosome 1 (1p copy loss and 1q copy gain) and chromosome 7 (7p11.2 focal amplification and 7q31.2 copy gain) are frequently detected in plasma cfDNA of patients that were resistant to EGFR-TKI (as with either intrinsic or acquired resistance). Other chromosomal genetic variations detected frequently in the plasma cfDNA of these patients include chromosome losses of chr19, chr9, chr3, chr20, chr15, 16p, 22q, 10q and 8p, and chromosomal gains of 8q. Of interest, data analysis further revealed the certain patterns of chromosomal somatic CNV that may correlate to the intrinsic or acquired resistance to EGFR-TKI treatment in lung cancer patients. For example, 7p gains detected in the plasma cfDNA achieve highest correlation with predictable intrinsic EGFR-TKI resistance (ROC =0.93).
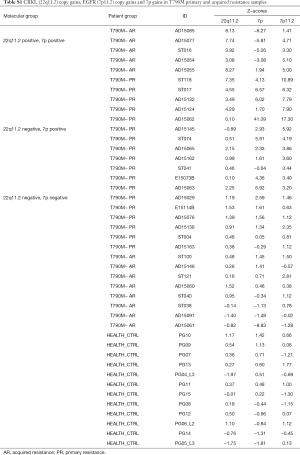
As regarding these chromosomal genetic variations, it is of interest that many lung cancer driver genes or tumor suppressor genes locate in the chromosome locus that have detectable changes in patients with lung cancer that are resistant to EGFR-TKI, as shown in this study. Of them, PCDHA gene cluster are the most commonly hyper-methylated genes discovered in human cancers (26,27). CRKL and EGFR are two well-studied lung cancer oncogenes: CRKL amplification was discovered as one of the acquired resistances to kinase inhibitors in lung cancers treated with EGFR inhibitors (25); EGFR amplification is one of the most common genetic events in lung cancer (33), and gain of EGFR amplifications has been proposed as one of the potential drug resistance mechanism of EGFR tyrosine kinase inhibitors (34,35). In our research, EGFR (7p11.2) copy gains were detected in 16/31 (51.6%) plasma samples. CRKL (22q11.2) copy gains were detected in 8/31 (25.8%) plasma samples. Of note, however, CRKL and EGFR copy gains were detected in separate samples (Table S1), it is thus suggested that these two genetic events, if they are corresponding to, are independent to EGFR-TKI resistance. On the other hand, our study also suggest that EGFR gene amplification can drive EGFR-TKI resistance, with gene amplification may results in a ligand independent kinase domain activation leading to intrinsic TKI resistance while secondary EGFR mutation is a result of genetic selection for gained resistance during TKI treatment.
Full table
TKI resistance remains as a clinical challenge for lung cancer management. In this study, we determined the chromosomal somatic copy number changes in plasma samples of patients with TKI resistance by using clinical achievable pipeline ultra-sensitive chromosomal aneuploid detector (UCAD). Our results not only identified the most frequently detectable chromosomal somatic copy number variants that are associated with TKI resistance, but also revealed patterns that may specifically correlate with either intrinsic or acquired EGFR-TKI resistance. Our data also demonstrated that some of the chromosomal somatic copy number variants such as 7p gain and 1q gain predict worse survival of the patients. These novel findings have significant clinic impacts for guiding lung cancer treatment. For examples, EGFR gene amplification (7p gain) in lung cancer cells may result in intrinsic resistance to TKI, and these patients may benefit from clinical management with addition of cetuximab, an anti-EGFR monoclonal antibody that target EGFR amplification; Chr1q gain is another frequent genetic event identified in lung cancer patients with intrinsic resistance to TKI, and 1q21.3-encoded S100 calcium-binding protein (S100A) family members and IL-1 receptor–associated kinase 1 (IRAK1) which can be targeted by a small-molecule kinase inhibitor, pacritinib; The examination of the potential 7p gains and 1q gain in the plasma samples of lung cancer patients with UCAD may provide a useful tool for monitoring EGFR-TKI response and drug resistance assessment in patient.
Conclusions
In this study, we identified multiple somatic CNV in distinguishing EGFR-TKI intrinsic and acquired resistance through plasma cfDNA sequencing. The results were from a small-scale prospective study, including 31 cancer patients and 10 health controls. The data present here uncovered encouraging findings for mechanism and biomarkers for EGFR-TKI resistance. However, a large prospective clinical trial to further confirm these discoveries is urgently needed.
Acknowledgments
Funding: This work was supported by grants from Projects of Science and Technology Project of Hangzhou Bureau (20170533B28), the Zhejiang Provincial Natural Science Foundation (LY19H160032 and LQ20H160020) and National Natural Science Foundation of China (81602671, 81602555 and 81773242), Major project of Hangzhou Science and Technology Bureau (20180417A01).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The protocol was approved by the Institutional Review Board of Hangzhou First People’s Hospital (No. HZFH2015-47-01). All patients signed the informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. [Crossref] [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637-58. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Gou LY, Li AN, Yang JJ, et al. The coexistence of MET over-expression and an EGFR T790M mutation is related to acquired resistance to EGFR tyrosine kinase inhibitors in advanced non-small cell lung cancer. Oncotarget 2016;7:51311-9. [Crossref] [PubMed]
- Carrera S, Buque A, Azkona E, et al. Epidermal growth factor receptor tyrosine-kinase inhibitor treatment resistance in non-small cell lung cancer: biological basis and therapeutic strategies. Clin Transl Oncol 2014;16:339-50. [Crossref] [PubMed]
- Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med 2015;372:1700-9. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Kuznetsova AY, Seget K, Moeller GK, et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 2015;14:2810-20. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Bakhoum SF, Landau DA. Chromosomal Instability as a Driver of Tumor Heterogeneity and Evolution. Cold Spring Harb Perspect Med 2017. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Zojer N, Dekan G, Ackermann J, et al. Aneuploidy of chromosome 7 can be detected in invasive lung cancer and associated premalignant lesions of the lung by fluorescence in situ hybridisation. Lung Cancer 2000;28:225-35. [Crossref] [PubMed]
- Ricke RM, van Deursen JM. Aneuploidy in health, disease, and aging. J Cell Biol 2013;201:11-21. [Crossref] [PubMed]
- Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med 2015;372:1589-97. [Crossref] [PubMed]
- Stover DG, Parsons HA, Ha G, et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J Clin Oncol 2018;36:543-53. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]
- Xia S, Huang CC, Le M, et al. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer 2015;90:78-84. [Crossref] [PubMed]
- Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010;28:357-60. [Crossref] [PubMed]
- Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065-9. [Crossref] [PubMed]
- Liang D, Lv W, Wang H, et al. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat Diagn 2013;33:409-15. [Crossref] [PubMed]
- Zhang S, Zhu L, Xia B, et al. Epidermal growth factor receptor (EGFR) T790M mutation identified in plasma indicates failure sites and predicts clinical prognosis in non-small cell lung cancer progression during first-generation tyrosine kinase inhibitor therapy: a prospective observational study. Cancer Commun (Lond) 2018;38:28. [Crossref] [PubMed]
- Suda K, Mizuuchi H, Murakami I, et al. CRKL amplification is rare as a mechanism for acquired resistance to kinase inhibitors in lung cancers with epidermal growth factor receptor mutation. Lung Cancer 2014;85:147-51. [Crossref] [PubMed]
- Jin H, Wang X, Ying J, et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene 2007;26:7490-8. [Crossref] [PubMed]
- El Hajj N, Dittrich M, Haaf T. Epigenetic dysregulation of protocadherins in human disease. Semin Cell Dev Biol 2017;69:172-82. [Crossref] [PubMed]
- Caballero OL, Cohen D, Liu Q, et al. Loss of chromosome arms 3p and 9p and inactivation of P16 (INK4a) in normal epithelium of patients with primary lung cancer. Genes Chromosomes Cancer 2001;32:119-25. [Crossref] [PubMed]
- Glukhova L, Lavialle C, Fauvet D, et al. Mapping of the 7q31 subregion common to the small chromosome 7 derivatives from two sporadic papillary renal cell carcinomas: increased copy number and overexpression of the MET proto-oncogene. Oncogene 2000;19:754-61. [Crossref] [PubMed]
- Fujisawa H, Reis RM, Nakamura M, et al. Loss of heterozygosity on chromosome 10 is more extensive in primary (de novo) than in secondary glioblastomas. Lab Invest 2000;80:65-72. [Crossref] [PubMed]
- Blaker H, von Herbay A, Penzel R, et al. Genetics of adenocarcinomas of the small intestine: frequent deletions at chromosome 18q and mutations of the SMAD4 gene. Oncogene 2002;21:158-64. [Crossref] [PubMed]
- Krijgsman O, Carvalho B, Meijer GA, et al. Focal chromosomal copy number aberrations in cancer-Needles in a genome haystack. Biochim Biophys Acta 2014;1843:2698-704.
- Levva S, Kotoula V, Kostopoulos I, et al. Prognostic Evaluation of Epidermal Growth Factor Receptor (EGFR) Genotype and Phenotype Parameters in Triple-negative Breast Cancers. Cancer Genomics Proteomics 2017;14:181-95. [Crossref] [PubMed]
- Tokumo M, Toyooka S, Ichihara S, et al. Double mutation and gene copy number of EGFR in gefitinib refractory non-small-cell lung cancer. Lung Cancer 2006;53:117-21. [Crossref] [PubMed]
- Nukaga S, Yasuda H, Tsuchihara K, et al. Amplification of EGFR Wild-Type Alleles in Non-Small Cell Lung Cancer Cells Confers Acquired Resistance to Mutation-Selective EGFR Tyrosine Kinase Inhibitors. Cancer Res 2017;77:2078-89. [Crossref] [PubMed]


