Long non-coding RNA growth arrest specific-5: a potential biomarker for early diagnosis of severe asthma
Introduction
Bronchial asthma is a chronic airway inflammation disease with heterogeneity in etiology, pathogenesis, clinical manifestations and prognosis. Severe asthma (SA) is a subset of asthma, which is the main reason for death and disability caused by asthma.
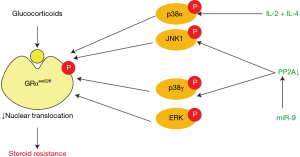
Glucocorticoid resistance is a key feature of SA. Glucocorticoids bind to and activate glucocorticoid receptor (GR) in cytoplasm. Activated cytosolic GR translocates to nucleus and combines with glucocorticoid response element (GRE), consequently promoting the transcription of anti-inflammatory genes and repressing the transcription of pro-inflammatory genes. So that interfering with the binding of GR to DNA can act as an inhibitor of steroid action (1). The molecular mechanisms that contribute to glucocorticoid resistance are complicated and have not been fully clarified, although several possible molecular mechanisms have been presented (2). Currently, glucocorticoid receptor phosphorylation at serine 226 (GRser226) is thought to play a vital role in glucocorticoid resistance. Over expression of interleukin (IL)-2 and IL-4 is found in the airways of patients with steroid-resistant asthma (3). IL-2 plus IL-4 in combination reduce GR nuclear translocation by phosphorylation of the GRαser226 through p38 mitogen-activated protein kinase α (p38MAPKα) pathway in T cells, which can be blocked by p38MAPK inhibitor (4-6). A large proportion of patients with SA demonstrate reduced GR nuclear translocation and GRE binding activity in peripheral blood monocular cells (PBMCs) after intervention with glucocorticoid, which can be explained by phosphorylation of GRαser226 (7). p38MAPK inhibitor inhibits GRser226, resulting in increased nuclear translocation (8). GR can also be phosphorylated directly by another MAPK, namely c-Jun N-terminal kinase (JNK), which is involved in GR phosphorylation in PBMCs from patients with SA (9). Moreover, a recent study has found the serine/threonine phosphatase protein phosphatase 2A (PP2A) can dephosphorylate GRser226 through JNK1 pathway in PBMCs from patients with SA (10).
A study revealed that miR-9 expression was increased in pulmonary macrophages and lung samples from interferon-γ (IFN-γ)/lipopolysaccharide (LPS)-induced steroid-resistant mouse model and in sputum samples from patients with neutrophilic asthma (11). MiR-9 decreased PP2A activity and dexamethasone-induced GR nuclear translocation through targeting protein phosphatase 2 regulatory subunit B δ isoform (PPP2R5D), a key regulator of PP2A. Decreased PP2A led to increased phosphorylation of p38MAPKγ and JNK1, and further led to an increased phosphorylation of GRser226 (11). But p38MAPKγ mainly exists in skeletal muscle tissue while lung tissue mainly expresses p38MAPKα (12,13), therefore, miR-9 regulates GR phosphorylation mainly from JNK1 pathway (Figure 1).
Long non-coding RNAs (lncRNAs) are a diverse class of RNA transcripts with a length of >200 nucleotides that do not encode proteins (14). LncRNAs are important regulators of gene expression and cellular function (15). Growth arrest specific-5 (GAS5) gene is a small nucleolar RNA host gene, containing multiple C/D box small nucleolar RNA genes in its introns (16). Part of its secondary RNA structure of the encoded transcript mimics GRE which means it can bind to DNA binding domain (DBD) of GR (17). This action blocks GR from being activated and thereby stops it from regulating the transcription of its target genes, thus implicating a potential promising role in the regulation of glucocorticoid sensitivity and resistance. There are two isoforms of GAS5, GAS5a and GAS5b, and the GAS5 GRE-mimic is located in exon 12, which is found in both isoforms (17). Human bronchial epithelial cells (HBECs), PBMCs, and mice lung tissue mainly express GAS5a (16), and the following GAS5 is all referring to GAS5a.
Finding a biomarker to evaluate the glucocorticoid responsiveness is extremely important for diagnosis and treatment of patients with SA. However, till now, few effective and sure-enough biomarkers to evaluate the glucocorticoid responsiveness of patients with asthma can be available. Based on above elaboration, the present study aimed to illuminate the role of GAS5 in patients with SA and to verify the possibility of GAS5 as a biomarker for diagnosis of SA.
Methods
Clinical subjects
Five adult patients with SA, twenty non-severe asthma (nSA) and twenty control subjects were recruited from the Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University. The diagnosis of SA was according to 2014 European Respiratory Society (ERS)/American Thoracic Society (ATS) guidelines (18), and the inclusion criteria were as follows: (I) FEV1 ≥40% predicted, and FEV1 ≥55% predicted after bronchodilator treatment; (II) less than 6 times of acute exacerbations in the last 6 months and no acute exacerbation in previous 1 month before grouping; (III) no hospitalization for asthma in the last 6 months; (IV) no tracheal intubation due to asthma in the last 1 year; (V) oral administration of prednisolone less than 20mg per day; (VI) non-smoking patients. Asthmatic patients not meeting the criteria of SA were considered as nSA. Control subjects were healthy volunteers who had no history of asthma, allergic and immune system diseases, and were non-smokers. Participants with cardiac insufficiency, abnormal liver function, pulmonary embolism, coinfection, tuberculosis, blood system diseases, malignant tumor, and other diseases were not included. This study was approved by the ethics institute of Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University and the signed informed consent was obtained from all participants (KY2018_016).
PBMCs separation and culture
PBMCs were enriched by Ficoll (GE Healthcare, Massachusetts, USA) gradient centrifugation within 4 hours. The cell viability determined by trypan blue exclusion assay was greater than 95%. PBMCs were washed twice with phosphate buffer saline (PBS, Beyotime Institute of Biotechnology, Shanghai, China), and then cultured in 6-well plates (1×104/well) at 37 °C, 5% CO2 in RPMI-1640 (Gibco, Invitrogen, UK) containing 10% foetal bovine serum and 1% penicillin, streptomycin. Twenty-four hours later, 100 nmol/L dexamethasone was added to each well and cells were incubated for another 24 hours.
Human bronchial epithelial (16HBE) cells culture, transfection and interoperations
16 HBE cells were purchased from Zhong Qiao Xin Zhou Biotechnology Co., Ltd (Shanghai, China). Cells were seeded on 6-well plates (1×104/well) in RPMI-1640 medium at 37 °C, 5% CO2 in a humidified incubator. Cells between passages 4 to 10 were used for all experiments. MiR-9 mimic and JNK1 inhibitor were transfected into 16HBE cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA), according to the kit instructions and incubated for 2 days at 37 °C or in the presence of IL-2 (50 IU/mL; R&D Systems, Abingdon, United Kingdom) and IL-4 (50 IU/mL; R&D Systems, Abingdon, United Kingdom), cells were cultured for 48 hours at 37 °C. All nucleic acids were purchased from GenePharma company (Shanghai, China) if not particularly indicated.
Establishment of mouse model of steroid-insensitive asthma
Twenty-four female BALB/c mice (aged 6 weeks) were purchased from Lingchang Biotechnology Co., Ltd (Shanghai, China), and were randomly and averagely divided into 3 groups, i.e., control group, asthma group and dexamethasone group. All animal studies were approved by the Institutional Animal Care and Use Committee of the Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University. A murine model of steroid-insensitive asthma was established in accordance with the method described by Komlósi et al. (19). Briefly, on days 1 and 14, the mice in asthma group and dexamethasone group were sensitized by intraperitoneal injection of 20 µg ovalbumin (OVA) (Grade V; Sigma, USA) emulsified in 2.25 mg aluminium hydroxide (Thermo Fisher Scientific, Waltham, MA, USA) in a total volume of 0.1 mL PBS. On days 28, 29 and 30, the mice were placed in a plastic chamber and challenged with aerosolized 1% OVA for 20 min per day. On day 27, the day before the first OVA challenge, the mice were instilled intranasally with 10 µg (60 µL) lipopolysaccharide (LPS) (Beyotime Institute of Biotechnology, Shanghai, China). On days 29 and 30, the mice in dexamethasone group were injected subcutaneously with dexamethasone (Fengyuan Inc., Ma’anshan, China), 5 mg/kg/day with a total volume of 0.1 mL. Other groups received 0.1 mL normal saline as placebo. The mice in control group received the same schedule for sensitization and challenge with an equivalent amount of PBS instead of OVA and LPS. On day 31, 24 h after last challenge, mice were sacrificed, and lung tissue samples were collected and preserved in liquid nitrogen.
Analysis of bronchoalveolar lavage fluid (BALF)
BALF were collected by lavaging the lungs 3 times with 0.5 mL of ice-cold 0.9% sodium chloride. The BALF was centrifuged (1,000 g, 15 min, 4 °C), and the supernatant were stored at −80 °C for further detection. The resulting pellets were resuspended in PBS for total inflammatory cell counts with a hemocytometer and a microscope. The levels of IL (interleukin)-4, IL-13 and IFN-γ (interferon-γ) in BALF were detected by enzyme-linked immunosorbent assay (MultiSciences, Hangzhou, China) according to the manufacturer’s instructions.
Lung histology
The left lung was obtained from the sacrificed mice and fixed in 10% neutral-buffered formalin. The lung was thoroughly dehydrated prior to embedding in paraffin. Paraffin sections (5 µm) were then stained with either hematoxylin-eosin (HE) or periodic acid-Schiff (PAS). To quantify airway global cells, we used a 5-point grading system: 0, no goblet cells; 1, <25% of the epithelium; 2, 25–50% of the epithelium; 3, 50–75% of the epithelium; 4, >75% of the epithelium. Scoring of goblet cells was performed in at least three different fields for each lung section.
RNA isolation and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted using a TRIzol reagent (Invitrogen, Carlsbad, USA), and first-strand cDNA was synthesized using first Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The expression of RNA was measured by qRT-PCR (ABI PRISM7500, USA) with AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China). Relative expression of GAS5 was normalized with U6. The comparative Ct (ΔΔCt) method was used for quantification of gene expression. The primers used for qRT-PCR were designed and synthesized by GenePharma Co., Ltd (Suzhou, China) (Table 1).

Full table
Protein extraction and Western blot
Total protein from PBMCs and 16HBE cells was obtained using Protein Extraction Kit (Beyotime Institute of Biotechnology, Shanghai, China) supplemented with a phosphatase inhibitor cocktail (Beyotime Institute of Biotechnology, Shanghai, China). The cell lysates were subjected to Western blot using the anti-phospho-glucocorticoid receptor (Ser226) antibody (Cell Signaling Technology, Boston, USA), anti-GAPDH antibody (Abcam, Cambridge, UK), and mouse anti-rabbit horseradish peroxidase-conjugated IgG (Santa Cruz Biotechnology). The intensity of each protein band was visualized by FlourChemQ system (Proteinsimple, CA, USA) and quantitated against the internal loading control GAPDH with ImageJ version 1.48 (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Differences between two groups were analyzed by t-test. Differences among more than two groups were assessed by one-way analysis of variance (ANOVA) followed by the Bonferroni multiple comparison tests, and differences between gender was assessed by Chi-square test. Data were expressed as mean ± standard error of the mean (SEM). A P values <0.05 was considered statistically significant. Statistical analysis was done by using SPSS software (version 21.0; IBM, Chicago, IL, USA).
Results
Characteristics of experimental subjects
As shown in Table 2, there were no statistically significant differences in age, gender, EOS, FeNO, BMI between SA and nSA group. Patients with SA had lower FEV1/FVC%, and FEV1% (of predicted) as compared to nSA.

Full table
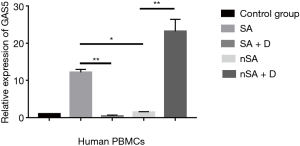
Levels of GAS5 and GRser226 in PBMCs before and after dexamethasone treatment in vitro
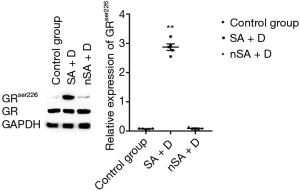
As shown in Figure 2, the level of GAS5 in the PBMCs from SA was higher than that in nSA (P<0.05). Surprisingly, after treatment with dexamethasone (100 nmol/L) for 24 h in vitro, the expression of GAS5 in SA decreased over 15-fold lower, while increased over 20-fold higher in nSA (P<0.001). Moreover, the expression of GRser226 in SA group was higher than that in nSA group after dexamethasone treatment (P<0.001, Figure 3).
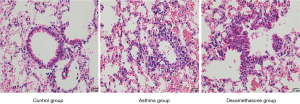
Comparison of IFN-γ, IL-4 and IL-13 in BALF of mouse model
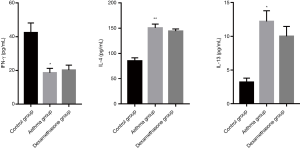
The expression of IFN-γ of asthma and dexamethasone group was lower than that of control group (P<0.05), conversely, the expression of IL-4 of asthma and dexamethasone group was higher than that of control group (P<0.001), and IL-13 showed similar trend (P<0.05), while there is no significant difference between asthma group and dexamethasone group in IFN-γ, IL-4 and IL-13 expression (Figure 4).
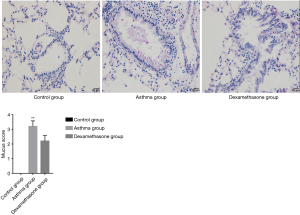
HE and PAS of lung tissue of mouse model
Asthma group showed an increased level of airway inflammation in lung tissue of mice in HE staining, while dexamethasone group non-significantly attenuated airway inflammation (Figure 5), and there is no significant difference between asthma group and dexamethasone group in mucus score (Figure 6).
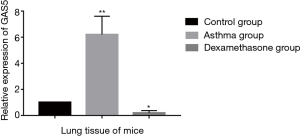
Expression of GAS5 in the lung tissue of mouse model
The expression of GAS5 in the lung tissue of murine model of steroid-insensitive asthma was higher significantly than that of controls (P<0.001, Figure 7). When treated with dexamethasone, the expression of GAS5 in steroid-insensitive asthmatic mice decreased dramatically (P<0.001), even lower than that in controls (P<0.05).
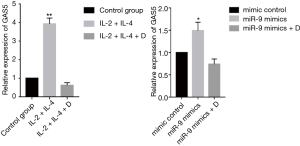
Expression of GAS5 and GRser226 in 16HBE cells
Treating 16HBE cells with IL-2 plus IL-4 increased the expression of GAS5 (P<0.001), but when adding dexamethasone, the expression of GAS5 decreased remarkablely (P<0.001, Figure 8). Similarly, the GAS5 level in 16HBE cells transfected with miR-9 mimics was also reversed by dexamethasone (P<0.05, Figure 8). Furthermore, both treatment with IL-2 plus IL-4 and transfection of miR-9 mimics could evaluate the GRser226 level in 16HBE cells (P<0.001, Figure 9).
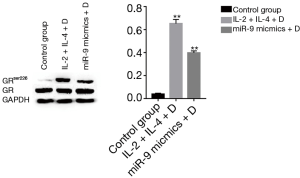
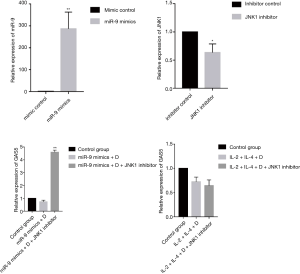
Role of JNK1 inhibitor in the expression of GAS5 in 16HBE cells
As shown in Figure 10, the expression of GAS5 in IL-2 + IL-4 + dexamethasone group was not affected by transfection of JNK1 inhibitor. However, transfection of JNK1 inhibitor could reverse the expression of GAS5 in miR-9 mimics + dexamethasone group (P<0.001).
Discussion
In the present study, we found an increased expression of GAS5 in the PBMCs from patients with SA when compared with that from patients with nSA. After treatment with dexamethasone in vitro, the expression of GAS5 was downregulated in SA, while it was upregulated in nSA. Moreover, the expression of GRser226 in SA was higher than that in nSA after dexamethasone treatment in vitro. In a mouse model of steroid-insensitive asthma, dexamethasone treatment also decreased the expression of GAS5, which was consistent with the results from PBMCs. Both treatment with IL-2 plus IL-4 and transfection of miR-9 mimics could increase the expression of GRser226, and the expression of GAS5 was downregulated by dexamethasone intervention. Transfection with JNK1 inhibitor could not affect the expression of GAS5 in IL-2 + IL-4 + dexamethasone treated 16HBE cells, but reverse the expression of GAS5 in miR-9 mimics + dexamethasone intervened 16HBE cells. To our knowledge, these results were the first time to demonstrate that, GAS5 was differently expressed between SA and nSA, and was affected by dexamethasone intervention, highlighting a potential role of GAS5 as a biomarker for diagnosis of SA.
GAS5 was first found in growth arrest cells when the concept of lncRNA is merging in 1988 to 1992 (20,21), but the researches that follows are mainly about the role of GAS5 acting as tumor suppressor of various kinds of tumors. Studies between GAS5 and asthma was first seen in 2018 (22), GAS5 promoted airway smooth muscle cell proliferation in asthma and knockdown of GAS5 decreased airway hyperresponsiveness in asthmatic rats. And several studies indicated that GAS5 associated with glucocorticoid resistance (17,23). Therefore, GAS5 could be seen an offensive factor in asthma. Growth arrest by serum starvation or treatment with inhibitors of protein translation is associated with attenuated translation of 5’TOP RNAs and the inhibition of their degradation, resulting in marked accumulation of spliced, mature GAS5 (17). We found dexamethasone could elevate the expression of GAS5 in PBMCs of patients with nSA, which was consistent with the findings in airway epithelial cells by Keenan et al. (24) and findings in nucleus of HeLa cells by Kino et al. (17). However, they did not illustrate the possible reason why dexamethasone could elevate the expression of GAS5. We thought it was attributed to glucocorticoid-induced reduced lifespan of various type of cells (25,26). Glucocorticoids affect cell survival decisions through causing apoptosis by themselves or through changing the vulnerability of cells to apoptotic stimuli (27). GAS5 is so named because it accumulated in growth-arrested cells, and it promotes apoptosis in many cells, e.g., cardiomyocyte (28), malignant melanoma cell (29), macrophage and vascular endothelial cell (30). It is possible that glucocorticoid induces apoptosis by upregulating GAS5, and the role of GAS5 playing in the mechanisms of glucocorticoid-induced apoptosis needs further research. On the contrary, Lucafo et al. reported that a slight reduction of GAS5 expression after treatment with methylprednisolone for 72h in PBMCs from healthy volunteers was observed (23). We hold the opinion that the treatment time of glucocorticoid might be the reason. In other words, the upregulation of GAS5 mediated by glucocorticoid might be transient and continuous glucocorticoid intervention could not keep GAS5 at high level. Further experiments should be carried out to clarify the effect of different treatment time of glucocorticoid on GAS5 expression.
It is worth noting that the expression of GAS5 in PBMCs from patients with SA was sharply decreased after dexamethasone treatment, considering the strong relationship between GAS5 and phosphorylation of GRser226, we guessed that high expression of GRser226 might be the reason for the low expression of GAS5 in SA after dexamethasone treatment. Failed translocation of GRser226 to nucleus could not elevate the expression of GAS5, but GAS5 could still bind to the DBD of GRser226, which resulted in a remarkable decrease of GAS5 level. To verity our speculation, we established a mouse model of steroid-insensitive asthma and found the similar sharp decrease in this model. To further verify the results in PBMCs from patients with SA, we used 16HBE cells as cell model to perform in vitro experiment. Both treated with IL-2 plus IL-4 and transfected with miR-9 mimics could elevate the expression of GRser226 in 16HBE cells, however, miR-9 mimics elevate the level of GRser226 through JNK1 pathway while IL-2 plus IL-4 elevate the expression of GRser226 through p38MAPKα pathway. On the foundation of elevated expression of GRser226, the GAS5 expression decreased much lower in 16 HBE cells after dexamethasone treatment, which meant we could duplicate the downregulation of GAS5 in SA after dexamethasone treatment by increasing the level of GRser226. In addition, the expression of GAS5 after blocking JNK1 pathway by JNK1 inhibitor was reversed in miR-9 mimics + dexamethasone group, but the level of GAS5 in IL-2 + IL-4 + Dexamethasone group was not affected by JNK1 inhibitor, which further verified our speculation. Overall, upregulation of GRser226 might be the reason for the low expression of GAS5 in SA after dexamethasone treatment.
The prevalence of SA was estimated between 5% and 10% of all asthmatic patients (31). Although the proportion is not high, SA accounts for huge medical expenses because of frequent acute exacerbation and emergency hospitalization (32). According to the ERS/ATS guidelines, SA is defined as asthma which requires high dose inhaled corticosteroids plus a second controller or treatment with systemic corticosteroids for ≥50% of the previous year to control, or which remains uncontrolled despite this therapy (18). The diagnosis of SA needs a medication history of 6 months or more. However, what clinical history provides is a vague impression on responsiveness to glucocorticoid of patients with SA, and is indistinct due to many other reasons such as treatment adherence, inhaler technique and coexisting conditions (33,34). It is very likely for clinicians to have cognitive bias towards glucocorticoid sensitiveness of patients with SA. Efforts have been made on finding biomarkers for the prognosis of SA (35). For example, when diagnosis of SA was confirmed, high level of FeNO could identify those with more severe symptoms and frequent exacerbation, but no difference was found in the FeNO level of SA as compared with nSA (36). Elevated sputum neutrophils with or without concurrent eosinophilia have been found to be associated with a more SA phenotype (37). In addition, elevated level of periostin, a matricellular protein which is secreted by bronchial epithelial cells and lung fibroblasts in response to Th2 cytokines, has been found to be associated with more severe and eosinophilic asthma (38). However, these biomarkers are all based on the confirmed diagnosis of SA and not efficient to distinguish SA from nSA. In this study, we found higher expression of GAS5 in SA than that in nSA. But the cutoff value of GAS5 was hard to be defined due to individual difference, and could not be used as a biomarker to distinguish SA from nSA efficiently. However, upregulation of GAS5 in nSA and downregulation in SA after dexamethasone treatment in vitro formed a sharp contrast, which could be used as a potential biomarker for diagnosis of SA.
Till now, there is no satisfactory way to quantify the sensitivity of glucocorticoid in patients with SA. It has reported that halving GAS5 level in airway epithelial cells by GAS5-targeting siRNA can double dexamethasone induced GRE activity (24), which suggests a direct quantitative relation between GAS5 level and GRE activity, and reflects the process of competitive binding to the DBD of GR by GAS5. So that GAS5 might be used as a marker to quantify the sensitivity of glucocorticoid. But glucocorticoid resistance can be caused by reduced recruitment and disfunction of histone deacetylase 2, which is a key factor of inflammatory gene repression by glucocorticoid after binding to GRE (2,39). Further experiments should be carried out to illuminate the complex mechanisms.
However, our study had limitations. First, the sample size of SA was relatively small, which may be prone to false negatives due to low statistical power or fortuitous false-positive results. Second, asthma is a chronic disease, acute phase such as animal experiment and in vitro cell experiment are not necessarily to fully reflect this process. Third, the distribution of GAS5 in lung tissue by immunostaining was not available.
Conclusions
Taken together, we conclude that GAS5 can be used as a potential biomarker for diagnosis of SA. In addition, our results suggest the different expression of GAS5 may be caused by phosphorylation of GR, which provides a theoretical basis for further quantification of glucocorticoid sensitivity.
Acknowledgments
Funding: This study was supported by Jiangsu Province “Six Big Talent Peak” Project (WSN092), Changzhou High-Level Medical Talents Training Project (2016CZLJ017), Jiangsu Province “333 Talents” Project (BRA2016119) and the Changzhou Sci & Tech Program (CJ20179031).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-213). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. N Engl J Med 2005;353:1711-23. [Crossref] [PubMed]
- Barnes PJ. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol 2013;131:636-45. [Crossref] [PubMed]
- Leung DY, Martin RJ, Szefler SJ, et al. Dysregulation of interleukin 4, interleukin 5, and interferon gamma gene expression in steroid-resistant asthma. J Exp Med 1995;181:33-40. [Crossref] [PubMed]
- Irusen E, Matthews JG, Takahashi A, et al. p38 Mitogen-activated protein kinase-induced glucocorticoid receptor phosphorylation reduces its activity: role in steroid-insensitive asthma. J Allergy Clin Immunol 2002;109:649-57. [Crossref] [PubMed]
- Sher ER, Leung DY, Surs W, et al. Steroid-resistant asthma. Cellular mechanisms contributing to inadequate response to glucocorticoid therapy. J Clin Invest 1994;93:33-9. [Crossref] [PubMed]
- Matthews JG, Ito K, Barnes PJ, et al. Defective glucocorticoid receptor nuclear translocation and altered histone acetylation patterns in glucocorticoid-resistant patients. J Allergy Clin Immunol 2004;113:1100-8. [Crossref] [PubMed]
- Szatmáry Z, Garabedian MJ, Vilcek J. Inhibition of glucocorticoid receptor-mediated transcriptional activation by p38 mitogen-activated protein (MAP) kinase. J Biol Chem 2004;279:43708-15. [Crossref] [PubMed]
- Mercado N, Hakim A, Kobayashi Y, et al. Restoration of corticosteroid sensitivity by p38 mitogen activated protein kinase inhibition in peripheral blood mononuclear cells from severe asthma. PLoS One 2012;7:e41582. [Crossref] [PubMed]
- Ismaili N, Garabedian MJ. Modulation of glucocorticoid receptor function via phosphorylation. Ann N Y Acad Sci 2004;1024:86-101. [Crossref] [PubMed]
- Kobayashi Y, Mercado N, Barnes PJ, et al. Defects of protein phosphatase 2A causes corticosteroid insensitivity in severe asthma. PLoS One 2011;6:e27627. [Crossref] [PubMed]
- Li JJ, Tay HL, Maltby S, et al. MicroRNA-9 regulates steroid-resistant airway hyperresponsiveness by reducing protein phosphatase 2A activity. J Allergy Clin Immunol 2015;136:462-73. [Crossref] [PubMed]
- Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal 2000;12:1-13. [Crossref] [PubMed]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002;298:1911-2. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [Crossref] [PubMed]
- Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 1998;18:6897-909. [Crossref] [PubMed]
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8. [Crossref] [PubMed]
- Chung KF, Wenzel SE, Brozek JL, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J 2014;43:343-73. [Crossref] [PubMed]
- Komlósi ZI, Pozsonyi E, Tábi T, et al. Lipopolysaccharide exposure makes allergic airway inflammation and hyper-responsiveness less responsive to dexamethasone and inhibition of iNOS. Clin Exp Allergy 2006;36:951-9. [Crossref] [PubMed]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988;54:787-93. [Crossref] [PubMed]
- Coccia EM, Cicala C, Charlesworth A, et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol 1992;12:3514-21. [Crossref] [PubMed]
- Zhang XY, Tang XY, Li N, et al. GAS5 promotes airway smooth muscle cell proliferation in asthma via controlling miR-10a/BDNF signaling pathway. Life Sci 2018;212:93-101. [Crossref] [PubMed]
- Lucafo M, De Iudicibus S, Di Silvestre A, et al. Long noncoding RNA GAS5: a novel marker involved in glucocorticoid response. Curr Mol Med 2015;15:94-9. [Crossref] [PubMed]
- Keenan CR, Schuliga MJ, Stewart AG. Pro-inflammatory mediators increase levels of the noncoding RNA GAS5 in airway smooth muscle and epithelial cells. Can J Physiol Pharmacol 2015;93:203-6. [Crossref] [PubMed]
- Banuelos J, Shin S, Cao Y, et al. BCL-2 protects human and mouse Th17 cells from glucocorticoid-induced apoptosis. Allergy 2016;71:640-50. [Crossref] [PubMed]
- Kay P, Schlossmacher G, Matthews L, et al. Loss of glucocorticoid receptor expression by DNA methylation prevents glucocorticoid induced apoptosis in human small cell lung cancer cells. PLoS One 2011;6:e24839. [Crossref] [PubMed]
- Frankfurt O, Rosen ST. Mechanisms of glucocorticoid-induced apoptosis in hematologic malignancies: updates. Curr Opin Oncol 2004;16:553-63. [Crossref] [PubMed]
- Hao S, Liu X, Sui X, et al. Long non-coding RNA GAS5 reduces cardiomyocyte apoptosis induced by MI through sema3a. Int J Biol Macromol 2018;120:371-7. [Crossref] [PubMed]
- Chen L, Yang H, Yi Z, et al. LncRNA GAS5 regulates redox balance and dysregulates the cell cycle and apoptosis in malignant melanoma cells. J Cancer Res Clin Oncol 2019;145:637-52. [Crossref] [PubMed]
- Chen L, Yang W, Guo Y, et al. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS One 2017;12:e0185406. [Crossref] [PubMed]
- Hekking PP, Wener RR, Amelink M, et al. The prevalence of severe refractory asthma. J Allergy Clin Immunol 2015;135:896-902. [Crossref] [PubMed]
- O'Neill S, Sweeney J, Patterson CC, et al. The cost of treating severe refractory asthma in the UK: an economic analysis from the British Thoracic Society Difficult Asthma Registry. Thorax 2015;70:376-8. [Crossref] [PubMed]
- Israel E, Reddel HK. Severe and Difficult-to-Treat Asthma in Adults. N Engl J Med 2017;377:965-76. [Crossref] [PubMed]
- Lommatzsch M, Virchow JC. Severe asthma: definition, diagnosis and treatment. Dtsch Arztebl Int 2014;111:847-55. [PubMed]
- Wan XC, Woodruff PG. Biomarkers in Severe Asthma. Immunol Allergy Clin North Am 2016;36:547-57. [Crossref] [PubMed]
- Dweik RA, Boggs PB, Erzurum SC, et al. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med 2011;184:602-15. [Crossref] [PubMed]
- Moore WC, Hastie AT, Li X, et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol 2014;133:1557-63.e5. [Crossref] [PubMed]
- Kim MA, Izuhara K, Ohta S, et al. Association of serum periostin with aspirin-exacerbated respiratory disease. Ann Allergy Asthma Immunol 2014;113:314-20. [Crossref] [PubMed]
- Adcock IM, Ito K, Barnes PJ. Histone deacetylation: an important mechanism in inflammatory lung diseases. COPD 2005;2:445-55. [Crossref] [PubMed]