
Correlation of genomic alterations and PD-L1 expression in thymoma
Introduction
Thymic epithelial tumors (TETs) include several anterior mediastinal malignant tumours: thymomas, thymic carcinomas and thymic neuroendocrine cancers. There is significant diversity in the biologic features and clinical course of TETs, and thus the WHO classification thymomas comprises five subcategories A, AB, B1, B2, B3 based on cell morphology, degree of atypia, and differentiation of epithelial cells and lymphocytes (1). Many attempts have been made to identify target genes for successful therapy of TETs, that as many other cancers arise from germline mutations and/or somatic mutation accumulation during a lifetime. Next generation sequencing (NGS) represents a huge advancement in diagnostics aimed for sequencing large number of samples covering whole genome, whole exome or targeting genes of interest enabling large datasets for various cancers (2). These new molecular technologies revealed that thymic neoplasms have the lowest tumor mutation burden among all adult malignant tumours with a different pattern of molecular aberrations in thymomas and thymic carcinomas.
As for the PD-L1 expression in tumor cells in thymoma and thymic carcinoma, it varies a lot in published studies, with findings of PD-L1 expression from 23% to 92% in thymoma and 36% to 100% in thymic carcinoma. When correlated PD-L1 expression with disease stage some controversial results were obtained, with no association with tumor stage in most studies. This is, at least in part, explained by the fact that different PD-L1 immunohistochemical tests were applied in each trial, with four different antibodies (SP142, SP263, 22C3, and 28-8), different definition of PD-L1 positivity and cutoff values throughout the studies as well.
Today, there is a huge interest in using genomic features to produce predictive genomic-based immunotherapy biomarkers, particularly since recent data suggest that certain tumor-specific genomic alterations, either individually or in combination, appear to influence immune checkpoint activity and better responses as the outcome, so as such in some cancer types they may complement existing biomarkers to improve the selection criteria for immunotherapy.
Genomic alterations in thymoma
Frequent choice for exploring genomic alterations has been targeted NGS which targets hotspot regions or specific genes of interest. Targeted high-throughput NGS provides deep insight into the specifically selected hotspot regions and offers high coverage, which is extremely important for rare variants detection in heterogeneous tumors or in low purity samples. Since thymoma is a rare tumour, few studies regarding high-throughput sequencing datasets have been published. However, advances in different molecular tests, particularly NGS methods have shown that thymic tumors have the lowest tumor mutation burden among all adult cancers with a different pattern of molecular aberrations in thymomas and thymic carcinomas, and only a few significant mutations pointing to distinct molecular subtypes. Mutations in general transcription factor IIi (GTF2I) are unique to TETs, at high frequency in thymoma, with most frequent specific missense point mutation, p.(Leu404His) in the GTF2I oncogene, that is so unlikely to be found in other malignant tumors (3). GTF2I oncogene is mutated in 76–83% of types A and AB thymomas, less frequent in other subtypes and only in 8% of thymic carcinomas (3,4). TETs with a GTF2I mutation display a less aggressive clinical behaviour and better survival rates than those without it, which can explain higher GTF2I mutation prevalence in the indolent types A and AB thymomas (3) while thymic carcinomas, on the other side, display a higher mutational burden, with frequent mutations of TP53 and epigenetic regulatory genes and loss of CDKN2A.
Still, early studies with genomic analysis on thymoma, although limited in number due to disease rarity, have revealed several genes related to this pathology, such as EGFR, HER2, KIT, KRAS, and T53 (5). Belani et al. conducted the study in which whole genome/exome approach was used, and the findings pointed that DNMT3A (p.G728D) and ASXL1 (p.E657fs) variants are involved in thymoma genesis (6). Petrini and colleagues, using whole genome and transcriptome sequencing identified one translocation t(11;X), copy number gain of chromosome 1q, 5, 7 and X and CN loss of 3p, 6, 11q42.2-qter and q13, 10 SNVs and 2 indels, suggesting the need for additional screening for better understanding of disease genetics (3). Enkner’s group analyzed two B3 thymoma using Ion AmpliSeq Cancer Hotspot Panel, found a mutation in noncoding regions of the SMARCB1 and STK11 gene and nonsynonymous HRAS mutation in A thymomas (7). They discovered nonsynonymous variants in ERBB4 gene (erb-b2 receptor tyrosine kinase 4, a member of the epidermal growth factor receptor, EGFR, subfamily), but in more aggressive TETs (7). Wheler et al. (2013) (8) performed molecular analyses in 21 tumour samples, out of which by NGS in seven patients and PCR-based assays in an additional six patients, and detected diverse actionable mutations: PIK3CA (1 of 12 tested; 8%); EGFR (1 of 13; 8%); RET (1 of 7; 14%); and AKT1 (1 of 7; 14%). Heterogeneity in actionable molecular aberrations was noted, suggesting that multi-assay molecular profiling and individualizing treatment merits investigation. Gökmen-Polar et al. (2013) (9) have reported on a gene signature in order to determine metastatic behaviour in thymomas, so qRT-PCR assay for 23 genes (19 test and 4 reference genes) was performed on multi-institutional archival thymomas (n=36). Based on gene expression levels, tumors were divided into classes 1 and 2, that corresponded to low or high likelihood for metastases, and a computed signature was developed. This nine-gene signature that can predict metastatic behavior of thymomas was validated. as well. Tiseo et al. (2017) (10) investigated the mutational status of druggable genes (EGFR, c-KIT, KRAS, BRAF, PDGFR-alpha and -beta, HER2 and c-MET) and the expression of ALK and PD-L1 in 112 consecutive cases of TETs, but no mutations were found and no ALK positive case has been observed in thymoma patients.
Some studies were conducted on tumor tissue samples originating from patients who had been treated with chemotherapy previously like one of Wang and colleagues (11) who used NGS to analyze 197 cancer associated genes in malignant thymic neoplasms obtained from 78 pretreated patients (31 thymomas, 47 thymic carcinomas) with advanced-stage TETs. Somatic mutations were found in 39 genes, in 62% thymic carcinomas and 13% thymomas. Recurrent mutations were evidenced in 15 genes including BAP1, BRCA2, CDKN2A, CYLD, DNMT3A, HRAS, KIT, SETD2, SMARCA4, TET2, and TP53. Nine (23%) of 39 mutated genes are responsible for epigenetic regulatory proteins that are engaged in chromatin remodeling, histone modification, and DNA methylation, while recurrent mutations in 7 of those 9 (BAP1, ASXL1, SETD2, SMARCA4, DNMT3A, TET2, and WT1) were found in 34% samples of thymic carcinoma but not in thymoma. Thymic carcinomas displayed much higher mutational burden than thymomas and had frequent mutations of TP53, unlike thymoma (11). Nonsynonymous mutations in TP53 gene, important for disease pathogenesis (5) were detected also in some recent studies, in highly aggressive forms of the disease (7), and TP53 together with BCOR were the most frequently mutated genes in TCA and B3 thymomas, respectively (12).
Recent comprehensive multi-platform genomic analyses of TETs have been performed by Radovich et al. (13) The Cancer Genome Atlas network (TCGA) investigated 117 TET samples (107 thymomas, 10 thymic carcinomas) originating from treatment-naïve, predominantly early-stage disease patients. The authors used whole-exome sequencing (WES), and additionally, they analyzed methylation status, microRNA profile, gene expression profile by RNAseqtechnology, and copy number variations in the same cohort of patients. Recurrent somatic mutations were noted in the gene unique for TETs, GTF2I that is marked as a thymoma-specific oncogene, HRAS, NRAS and TP53, while four different molecular subtypes of TETs were identified that showed clinical and pathologic similarities to WHO subtypes B, thymic carcinoma, type AB and a mix of type A and AB (14).
Mutations in the thymoma-specific oncogene GTF2I were noted mostly in type A and AB thymoma, in line with data reported earlier (3). A significantly higher prevalence of aneuploidy was noted in thymomas originating from subpopulation with thymoma-associated myasthenia gravis (TAMG), confirming that the association of thymomas with myasthenia gravis (MG) is linked to an increased gene copy number variation, while gene expression profiling identified overexpressed autoantigens. This comprehensive work combined several approaches, including structural and functional analyses, in order to identified genomic events that underline TET pathogenesis.
So, irrespective of the GFT2I mutation, overall the lowest tumor mutational burden (TMB) of TETs among 21 other malignancies that were sequenced by TCGA has been confirmed (13). Thus, apart from the most frequent GTF2I, other genomic alterations in a cohort of 155 reported cases from several important studies (3,13-15), were noted by frequency as follows: HRAS, TP53, CYLD, PCLO, HDAC4, BCOR, PBRM1, BRD4, CSF1R, FGF3, NRAS, PAX7, PTPRB, ZMYM3, NOD1. However, in addition to GTF2I only a few other genes were recurrently mutated in TETs at a frequency of at least 3% in this cohort, HRAS, TP53, CYLD, PCLO and HDAC4. HRAS mutations affected types A and AB thymomas in ten of eleven mutated TETs. Thus, HRAS was the second most frequently mutated gene in thymomas in this cohort. TP53 and CYLD mutations do occur in thymomas but are more frequent in thymic carcinomas (8,11).
Lee and colleagues (16) based on findings on DNA mutations, mRNA expression and somatic copy number alterations from the TCGA TET data set, identified four molecular subgroups: tumours with GTF2I mutations, without GTF2I mutations but with expression of genes associated with T-cell signaling, and tumors with chromosomal stability and instability. These molecular subgroups corresponded with WHO subtypes A or AB, B1 or B2, B2, and B2, B3 or C, respectively. In one of the latest studies of thymoma transcriptomics, gene expression of 900 genes were analyzed in 31 thymoma patients using CapitalBioRNA microarray (17). It was demonstrated that 4 genes, E2F2, EPHA1, CCL25 and MCM2, were upregulated, while IL6, FABP4, CD36 and MYOC were downregulated. The emphasis of this study was on the expression level of genes involved in thymoma genesis (17). Recently, Yamaguchi et al. (18) performed NGS analysis of extracted DNA from fresh frozen surgically resected tissues (tumors and paired normal tissues) in 24 patients. DNA amplicon sequencing was performed with a custom panel of 53 cancer-related genes based on Ion AmpliSeqTM Cancer Hotspot Panel v2 comprising major oncogenes and tumor suppressor genes (including GTF2I). Unlike other studies findings, no genetic alterations were detected in 19 out of 24 patients. The nonsynonymous mutations of RAS gene which is known to have a significant role in pathogenesis of various malignant diseases, HRAS and NRAS (HRAS Q61R, HRAS G13R, and NRAS Q61K) was detected in three patients, and low frequently DNMT3A mutation was found in the other two patients (18).
Numerous differences have been recognized between TAMG and non-MG thymoma (NMG), but not at the molecular level, so Xi and colleagues (19) explored the differentially expressed genes between these two subtypes in order to reveal the molecular mechanisms important for the pathogenesis of TAMG. A significant difference between these two groups was evidenced regarding the expression level of 169 genes, with 94 up-regulated and 75 down-regulated genes. Overexpression of six genes in T cells (ATM, SFTPB, ANKRD55, BTLA, CCR7, TNFRSF25) important for the pathogenesis of TAMG and directly associated with autoimmune disease was detected. The overexpression of soluble BTLA (sBTLA) (P=0.027), CCR7 (P=0.0018), TNFRSF25 (P=0.0013) and ANKRD55 (P=0.0026) was identified and validated (19).
NGS analyzing 1,225×106 bp sequence from 35 thymoma patients with TruSeq Cancer Panel (TSACP) used for somatic variant detection in specific genomic regions, including 212 amplicons in 48 important cancer-related genes, revealed 1,963 potentially protein modulating variants including nonsense (N), frameshift (F), and missense (M) changes (20) Four genes, APC, ATM, ERBB4, and SMAD4, were the highest mutated genes having more than 100 NFM (nonsense, frameshift and missense) protein-changing variants, present in more than 70% of analyzed cases, pointing to their potential role in thymoma pathogenesis. Additionally, EGFR, FBXW7, FGFR3, FGFR2, GNAQ, GNA11, HNF1A, KIT, MET, PIK3CA, PTEN, and RB1 were highly mutated harboring more than 40 NFM changes. TP53 and KDR contained more than 90 NFM variants, out of which the majority were well known polymorphisms (familiar one rs1042522, and rs1870377) (20). Analyzing genetic findings and clinical data, they found that only the presence of variants in SMAD4 gene predicted significantly shorter overall survival. Recurrent mutations in this gene previously have been already discovered in other tumors with poor prognosis (12). As for APC gene, a key tumor suppressor factor involved in several fundamental cellular processes including tumorigenesis and homeostasis, especially of epithelial cells and lymphocytes (21), in this study found mutated in 27 patients, its alterations were associated with the aggressive subtypes B2 and B3, while “High risk alteration” at APC locus was noted in AB type that are not aggressive forms of thymoma, suggesting there is another tumor suppressor gene that have the opposite effect to APC (22). ATM gene encoding for the phosphatidylinositol 3-kinase, crucial for the repair of double-stranded DNA breaks, one of the most aberrant gene in solid and hematologic tumors, has been mutated in 26 out of 35 thymoma patients. Additionally, 168 recurrent variants were detected. On the other hand, some of the genes selected using TSACP panel including FGFR1, MPL, NPM, and SRC had less than 5 NFM variants (20). Peric et al. (20) found 24 out of 35 patients having in total 14 different nonsynonymous (NM) variants in ERBB4 gene, while Enkner and colleagues discovered nonsynonymous variants in ERBB4 gene only in more aggressive thymic epithelial tumors (TETs) (7). They also identified 2 TP53 variants with stop codon producing truncated protein with probably damaging function, and 4 missense variants (with one polymorphism) (20), compared to previous findings of nonsynonymous mutations in TP53 gene detected predominantly in highly aggressive forms of the thymic malignancies and infrequently in thymomas (7,11), and TP53 together with BCOR as the most frequently mutated genes in TCA and B3 thymomas, respectively (12). Additionally, they discovered KDR gene, encoding for tyrosine protein kinase, having variants in 74% cases, and PTEN gene, exhibiting missense variants with various oncogenic level in 71% of thymoma cohort (20) (Table 1).

Full table
Another concern also is identification and distinction of rare driver variants that cause disease development from passenger’s mutations, which have no influence on tumor phenotype. Somatic changes at DNA level in thymoma tissue represents a unique profile of a tumor in real-time, enabling personalized therapeutic approach (20), unlike some studies which emphasis was on the expression level of genes involved in thymoma genesis (17). Amplicon based technology such as Peric et al. (20) used for thymoma analyses, provides more reliable and usable data for optimal treatment options, due to high-coverage detection of low-frequency somatic variants (average coverage 145×), compared to lower coverage of whole-genome sequencing or WES analyses that was used in the work of Radovich et al. (13). Moreover, their investigation was more focused on concrete approach, analyzing disease-causing variants in hotspot regions of the most common mutated oncogenes, that could be responsible for disease origin or progression. Advances in targeted NGS enable great potential to analyze single driver variants and concurrent variants in different genes that lead to better understanding of the disease, and discovery of genetic markers that could be used for molecular-targeted therapeutics. Additionally, small targeted panels with high coverage, such as TSACP, have already been widely used for translational research, molecular diagnostics such as EGFR mutation in lung cancer (23).
Moreover, nowadays in the era of immunotherapy and imperfection of PD-L1 expression score as a biomarker criteria for immunotherapy, there is a huge interest in using genomic features to produce predictive genomic-based immunotherapy biomarkers, particularly since recent data suggest that certain tumor-specific genomic alterations, either alone or combined, appear to influence immune checkpoint activity and better responses of longer duration as the outcome. Thus, it might be expected that in some cancer types they may complement already established biomarkers in order to produce better selection criteria for immunotherapy.
Mechanisms of PD-L1 activation
The regulation of PD-L1 expression is essentially multilevel and complicated, differs between diverse tumor types and involves genetic, transcriptional and post-transcriptional pathways. PD-L1 and PD-L2 are encoded by the CD274 and PDCD1LG2 genes, respectively, that are integral parts of chromosome 9p.24.1, while PD-1 is encoded by the PDCD1 gene on chromosome 2q37.3 (24,25). The genomic alterations of the PD-L1/PD-L2 gene loci appear to be mainly responsible for PD-L1 expression both in malignant diseases.
PD-1/PD-L1 axis has an essential role in directing anti-tumour T-cell immune response and thus its regulation, the PD-1/PD-L1 interaction preventing the immune response against cancer. Binding of PD-1 with its ligands inhibits T-cell activation and anti-tumour activity.
Mechanisms of PD-L1 activation in cancer include a diversity of different processes: genomic alterations (copy number amplification, 3'-UTR disruption and other), constitutive oncogenic pathways activation, distinct extrinsic regulators (including interferon-g, inflammatory cytokines such as IL-17 and TNF-a, TGF-b1, and HIF-1a and epigenetic mechanisms, such as upregulation of oncogenic microRNAs (miRNAs), downregulation of tumor suppressor miRNAs, aberrant DNA methylation, and histone modifications (26) (Figure 1).
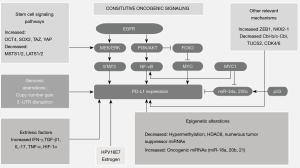
Transcriptional and posttranscriptional control of PD-L1 in cancer thus happens with variety of mechanisms included, and a number of signaling pathways are engaged, RAS/RAF/MEK/MAPK-ERK and PI3K/PTEN/Akt/mTOR. The activation of those pathways can be induced by oncogene mutations and/or by tumor suppressor genes alterations leading to loss of function. This results in two possible ways: direct action on target genes or the activation of transcription factors like STAT3, STAT1, c-Jun, HIFs, or NF-κB which inside the nucleus links to particular sites on PD-L1 gene promoter inducing its expression. PD-L1 is also directed post-transcriptionally by microRNAs, that links to mRNA resulting in its suppression or enhancement (24,26).
Activation of PD-L1 signaling pathway in the context of constitutive oncogenic signaling activation includes loss of PTEN expression, activation of different pathways including PI3K/AKT, RAS/MAPK, RAS/ERK/EMT and MAPK/ERK, inhibition of p53 signaling, upregulation of reprogramming factors (Oct4, Sox2, and c-Myc) and upregulation of ZEB1 [an inducer of epithelial-to-mesenchymal transition (EMT)] (26-28). Regulation of PD-L1 expression thus is directed via the PI3K/AKT and/or RAS/MAPK pathways in variety of cancer cell types. PD-L1 expression is repressed by the tumor suppressor gene PTEN which decreases PD-L1 expression while deletion of PTEN gene enhances PD-L1 expression via activating the PI3K/AKT signaling pathway (26,29-31). Recent findings point to a specific tumor-intrinsic function of PD-L1 in cancer development by orchestrating EMT, cancer stem cell -like phenotype, metastasis and resistance to therapy. There are emerging data on this tumor-intrinsic activity of PD-L1 in fostering malignancy development, metastasis, and resistance to therapy (26,32) (Figure 2).
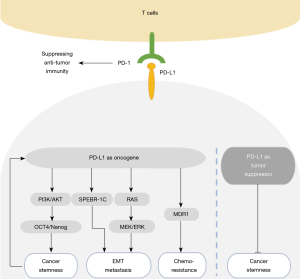
The expression of PD-L1 on cancer cells has been established as a biomarker used to select patients who will benefit from immunotherapy, but it is well recognized that clinically better outcome has also been observed in subpopulation with low PD-L1 expression.
PD-L1 expression in tumor cells of in thymoma and thymic carcinoma varies in published studies, with findings of PD-L1 expression from 23% to 92% in thymoma and 36% to 100% in thymic carcinoma. When correlated PD-L1 expression with disease stage, some controversial results were obtained among studies, with no association with tumor stage in most studies. This is, at least in part, explained by the fact that several diverse PD-L1 immunohistochemical tests were used in each trial, with four different antibodies (SP142, SP263, 22C3, and 28-8), different definition of PD-L1 positivity and cutoff values throughout the studies as well, so thus discrepancy has been evidenced in 47% of cases regarding PD-L1 expression levels (33,34). Moreover, as for PD-L1 expression there is intra- and inter-tumoral heterogeneity as well, and it also can vary over time, and a prominent problem with a diversity of the sensitivity and specificity of different IHC-based biomarkers, with the variable scoring thresholds applied.
Genomic-based immunotherapy biomarkers
Given all these facts, genomic-based biomarkers would appear useful to enable an alternative or complementary way to select those patients who may benefit from immunotherapy or be refractory to it. That is why a major interest arise in using genomic data to establish predictive immunotherapy biomarkers (genomic-based immunotherapy signatures), particularly since recent data suggest that certain tumor-specific genomic alterations, either individually or in combination, appear to influence immune checkpoint activity and better responses as the outcome, so as such in some cancer types they may complement existing biomarkers to improve the selection criteria for defining patients that would have benefit from immunotherapy.
There are examples of such tumor-specific genomic lesions like in triple negative breast cancer, with increased PD-L1 expression on tumor cells been linked to high mutation burdens, the total burden of copy number alterations in aneuploid tumors, to microsatellite instability (MSI), and to specific genomic driver alterations, including loss of tumor suppressor genes (PTEN), and activating mutations in driver oncogenes such as KRAS, EGFR and PIK3CA, BRCA mutant and BRCA-like HRD genomes (35). Similarly, deleterious gene mutational profiles in non-small cell lung cancer patient exomes were detected and based on these tumor genomics influence on cell signaling, PD-L1 expression, chemokines and immunosuppressive molecules, expression profiles of 24 chemokines and immunosuppressive molecules were explored in addition to PD-L1 expression in order to identify patients who would respond to PD-1 immunotherapy. The results of this study pointed that chemokine and immunosuppressive molecule expression profiles can be used for prediction of response to immunotherapy (36).
Thymoma-specific genomic lesions were investigated in last few years as already mentioned, but data on a correlation with PD-L1 expression are scarce.
Tiseo et al. (2017). (10) investigated the mutational status of druggable genes (EGFR, c-KIT, KRAS, BRAF, PDGFR-alpha and -beta, HER2 and c-MET) and the PD-L1 expression in 112 consecutive cases of TETs, but no mutation was detected, while PD-L1 expression was positive in 18% of thymomas, and high PD-L1 expression correlated with WHO classification stage type C (P<0.001) and Masaoka stage III–IV disease (P=0.007).
In the series of 35 thymoma tumor samples, nearly all obtained by surgery in stage I and II thymoma patients—all Caucasian population, PD-L1 expression using the clone 22c3 (Dako) was evidenced in 20 of them (57.1%), with high PD-L1 expression ≥50% in 8 (22.9%), and statistically significantly more PD-L1 expressors were in B2 thymoma cases. Significantly better survival was observed in PD-L1 negative cases. Great majority had PD-L1+/CD8+ subtype, but no significant difference in survival regarding PD-L1/CD8 subtypes as well as regarding histologic type was found. When PD-L1 expression and PD-L1/CD8 subtypes were correlated with NGS evidenced most frequent genomic alterations (APC, ATM, ERBB4, SMAD4, TP53, ALK, EGFR, KRAS, KDR, MET, PIK3CA, PTEN, RB1), significant differences were observed in the frequency of PD-L1 expression only in those with TP53 alterations, 24/35 cases (P=0.047), as well as in those with PTEN alterations, 24/35 cases (P=0.021), with no high PD-L1 expressors ≥50% among those without TP53 and PTEN alterations respectively. There was no correlation between PD-L1 expression and the number of NFM protein changing mutations (≥40%, ≥50%, ≥100%).
All these findings imply the complexity of genetics, distinct signaling pathways and pathogenesis of thymic tumours.
Conclusions
Advances in molecular technologies enabled genomic profiling of thymic tumors that have the lowest tumor mutation burden among all adult cancers, and detected distinct molecular subtypes. Although mutations in GTF2I are unique to TETs, the rarity of actionable mutations represents the big challenge for the development of biologic therapies. Recent research findings, despite of the diversity and infrequency of recurrent genetic alterations in thymoma, imply the need for further work to uncover druggable genomic targets and develop novel targeted drugs.
Genomic-based biomarkers would appear useful to enable an alternative or complementary way to select those patients who may benefit from immunotherapy or be refractory to it, particularly since recent data suggest that certain tumor-specific genomic alterations, either individually or in combination, appear to influence immune checkpoint activity and better and longer duration responses as the outcome. Thus, they may complement existing biomarkers to improve the selection criteria for defining patients that would have benefit from emerging immunotherapies.
This is of crucial importance since persistent autoreactive T cells in thymoma significantly elevate the risk for development of serious immune-related adverse events and thus decrease opportunities for use of immunotherapy.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Dragana Jovanovic and Semra Bilaceroglu) for the series “Thymoma” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/jtd-2019-thym-13). The series “Thymoma” was commissioned by the editorial office without any funding or sponsorship. DJ served as an unpaid Guest Editor of the series and serves as an unpaid editorial board member of Journal of Thoracic Disease from Feb 2019 to Jan 2021. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Okumura M, Ohta M, Tateyama H, et al. The World Health Organization histologic classification system reflects the oncologic behavior of thymoma: a clinical study of 273 patients. Cancer 2002;94:624-32. [Crossref] [PubMed]
- Raphael BJ, Dobson JR, Oesper L, et al. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Med 2014;6:5. [Crossref] [PubMed]
- Petrini I, Rajan A, Pham T, et al. Whole genome and transcriptome sequencing of a B3 thymoma. PLoS One 2013;8:e60572. [Crossref] [PubMed]
- Feng Y, Lei Y, Wu X, et al. GTF2I mutation frequently occurs in more indolent thymic epithelial tumors and predicts better prognosis. Lung Cancer 2017;110:48-52. [Crossref] [PubMed]
- Okuda K, Moriyama S, Haneda H, et al. Specific mutations in thymic epithelial tumors. Mediastinum 2017;1:16. [Crossref]
- Belani R, Oliveira G, Erikson GA, et al. ASXL1 and DNMT3A mutation in a cytogenetically normal B thymoma. Oncogenesis 2014;3:e111. [Crossref] [PubMed]
- Enkner F, Pichlhofer B, Zaharie AT, et al. Molecular Profiling of Thymoma and Thymic Carcinoma: Genetic Differences and Potential Novel Therapeutic Targets. Pathol Oncol Res 2017;23:551-64. [Crossref] [PubMed]
- Wheler J, Hong D, Swisher SG, et al. Thymoma patients treated in a phase I clinic at MD Anderson Cancer Center: responses to mTOR inhibitors and molecular analyses. Oncotarget 2013;4:890-8. [Crossref] [PubMed]
- Gökmen-Polar Y, Cook RW, Goswami CP, et al. A Gene Signature to Determine Metastatic Behavior in Thymomas. PloS One 2013;8:e66047. [Crossref] [PubMed]
- Tiseo M, Damato A, Longo L, et al. Analysis of a panel of druggable gene mutations and of ALK and PD-L1 expression in a series of thymic epithelial tumors (TETs). Lung Cancer 2017;104:24-30. [Crossref] [PubMed]
- Wang Y, Thomas A, Lau C, et al. Mutations of epigenetic regulatory genes are common in thymic carcinomas. Sci Rep 2014;4:7336. [Crossref] [PubMed]
- Moreira AL, Won HH, McMillan R, et al. Massively parallel sequencing identifies recurrent mutations in TP53 in thymic carcinoma associated with poor prognosis. J Thorac Oncol 2015;10:373-80. [Crossref] [PubMed]
- Radovich M, Pickering CR, Felau I, et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell 2018;33:244-258.e10. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform For exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Lee HS, Jang HJ, Shah R, et al. Genomic Analysis of Thymic Epithelial Tumors Identifies Novel Subtypes Associated with Distinct Clinical Features. Clin Cancer Res 2017;23:4855-64. [Crossref] [PubMed]
- Yu L, Ke J, Du X, et al. Genetic characterization of thymoma. Sci Rep 2019;9:2369. [Crossref] [PubMed]
- Yamaguchi H, Gyotoku H, Taniguchi H, et al. Genetic analysis of thymoma and thymic carcinoma [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2019; 2019 Mar 29-Apr 3; Atlanta, GA. Philadelphia (PA): AACR; Cancer Res 2019;79:abstract nr 1705.
- Xi J, Wang L, Yan C, et al. The Cancer Genome Atlas dataset-based analysis of aberrantly expressed genes by GeneAnalytics in thymoma associated myasthenia gravis: focusing on T cells. J Thorac Dis 2019;11:2315-23. [Crossref] [PubMed]
- Peric J, Samaradzic N, Trifunovic VS, et al. Genomic profiling of thymoma using targeted high-throughput approach. Archives of Medical Science 2020. [Crossref]
- Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci 2007;120:3327-35. [Crossref] [PubMed]
- Ströbel P, Hohenberger P, Marx A. Thymoma and thymic carcinoma: molecular pathology and targeted therapy. J Thorac Oncol 2010;5:S286-90. [Crossref] [PubMed]
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for NSCLC V.7.2019. ©National Comprehensive Cancer Network, Inc. 2019.
- Zerdes I, Matikas A, Bergh J, et al. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 2018;37:4639-61. [Crossref] [PubMed]
- Keir ME, Butte MJ, Freeman GJ, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677-704. [Crossref] [PubMed]
- Dong P, Xiong Y, Yue J, et al. Tumor-Intrinsic PD-L1 Signaling in Cancer Initiation, Development and Treatment: Beyond Immune Evasion. Front Oncol 2018;8:386. [Crossref] [PubMed]
- Chen J, Jiang CC, Jin L, et al. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol 2016;27:409-16. [Crossref] [PubMed]
- Mamessier E, Birnbaum DJ, Finetti P, et al. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann Transl Med 2018;6:54. [Crossref] [PubMed]
- Okita R, Maeda A, Shimizu K, et al. PD-L1 overexpression is partially regulated by EGFR/HER2 signaling and associated with poor prognosis in patients with non-small-cell lung cancer. Cancer Immunol Immunother 2017;66:865-76. [Crossref] [PubMed]
- Chen N, Fang W, Lin Z, et al. KRAS mutation induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother 2017;66:1175-87. [Crossref] [PubMed]
- Coelho MA, de Carné Trécesson S, Rana S, et al. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity 2017;47:1083-99.e6. [Crossref] [PubMed]
- Wang Y, Wang H, Yao H, et al. Regulation of PD-L1: Emerging Routes for Targeting Tumor Immune Evasion. Front Pharmacol 2018;9:536. [Crossref] [PubMed]
- Sekine I, Aida Y, Suzuki H. Expression patterns and prognostic value of PD-L1 and PD-1 in thymoma and thymic carcinoma. Mediastinum 2018;2:54. [Crossref]
- Sakane T, Murase T, Okuda K, et al. A comparative study of PD-L1 immunohistochemical assays with four reliable antibodies in thymic carcinoma. Oncotarget 2018;9:6993-7009. [Crossref] [PubMed]
- Barrett MT, Lenkiewicz E, Malasi S, et al. The association of genomic lesions and PD-1/PD-L1 expression in resected triple-negative breast cancers. Breast Cancer Res 2018;20:71. [Crossref] [PubMed]
- Brogden KA, Parashar D, Hallier AR, et al. Genomics of NSCLC patients both affirm PD-L1 expression and predict their clinical responses to anti-PD-1 immunotherapy. BMC Cancer 2018;18:225. [Crossref] [PubMed]


