
The impacts of ubiquilin 1 (UBQLN1) knockdown on cells viability, proliferation, and apoptosis are mediated by p53 in A549 lung cancer cells
Introduction
Ubiquilin (UBQLN) is a family of ubiquitin-like proteins which is composed of at least five members UBQLN1, UBQLN2, UBQLN3, UBQLN4, and UBQLNL. Among them, UBQLN1 is widely expressed in the plasma membrane, endoplasmic reticulum (ER), nucleus, cytoplasm, and autophagosomes in various tissues and cells. UBQLN1 plays an important role in maintaining protein balance through regulating the ubiquitin-proteasome system (UPS), macroautophagy (autophagy), ER-associated protein degradation (ERAD), and other signaling pathways (1,2). Furthermore, UBQLN1 can bind to various intracellular proteins, such as the proteasome and ubiquitin ligase (E3 ligase) (3), mammalian target of rapamycin (mTOR) (4), insulin-like growth factor receptor and insulin receptor (5) and etc., and therefore participates in the regulation of a variety of physiological or pathophysiological phenomena.
Current studies have evidenced that the expression level of UBQLN1 protein is closely associated with the occurrence, development, and clinical manifestations of some neurodegenerative diseases such as Alzheimer’s disease and Huntington’s disease (6,7). UBQLN1 protects mice from oxidative stress- and ischemia-induced neuronal damage by promoting the elimination of damaged proteins (8). Deficiency of UBQLN1 can cause ubiquitin-proteasome coupling disorders leading to myocardial ischemia-reperfusion injury in mice (9). It is noteworthy that UBQLN1 has also been implicated in tumorigenesis and cancer progression. UBQLN1 is low-expressed in human lung cancer tissues (10). Loss of UBQLN1 markedly enhances the metastatic properties and epithelial-to-mesenchymal transition (EMT) of human lung adenocarcinoma cells (10,11). In contrast, UBQLN1 is expressed at high levels in invasive breast cancer tissues and gastric cancer tissues, and is positively correlated with TNM stage, tumor invasion, and lymph node metastasis (12,13). In addition, UBQLN1 depletion has been found to increase the sensitivity of breast cancer cells to radiation therapy (14). All these findings strongly indicate that UBQLN1 plays a key role in the regulation of cancer development and progression. However, the underlying mechanism remained poorly understood.
p53 (also known as TP53) is a classic tumor suppressor gene involved in the regulation of various cellular processes in the cancer cell properties, such as apoptosis, proliferation, cell cycle arrest, or senescence (15,16). Mutant or deficiency of p53 has been found in more than half of all cancers including human non-small cell lung cancer (NSCLC) (15,17). However, the functional relationship between p53 and UBQLN1 remains ill-defined.
In the present study, we found that UBQLN1 knockdown (KD) resulted in a reduction of p53 protein levels in A549 cells by an autophagy-dependent mechanism. This finding deepens our understanding of UBQLN1 function in NSCLC and also suggests that UBQLN1 might be a promising target for the treatment of lung cancer.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-1362).
Methods
Antibodies and reagents
Antibodies against UBQLN1 (ab3341), p53 (ab32389), PERK (phosphor T982, ab192591), and IRE1 (phosphor S724, ab226974) were obtained from Abcam (Cambridge, UK). Anti-PERK (#3192), IRE1a (#3294), CHOP (#5554), Bax (#2772), Bcl-2 (#3498), Caspase-3 (#9662), mTOR (#2972), phosphor-mTOR (#2974), S6K (#9202), and phosphor-S6K (#9206) antibodies were purchased from Cell Signaling Technology ((Beverly, MA, USA). Anti-tubulin antibody (SAB4200715) and conventional reagents were purchased from Sigma-Aldrich (Boston, USA).
Cell culture
Human non-small cell lung carcinoma cell line A549 (ATCC® CCL-185TM) and H358 (ATCC® CRL-5807TM), and human normal lung epithelial BEAS-2B cells (ATCC® CRL-9609TM) were purchased from American Type Culture Collection (Rockville, MD, USA). Cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Sigma) at 37 °C in a humidified atmosphere containing 5% CO2.
Adenovirus construction and virus infection
Adenoviruses carrying human wild-type p53 cDNA and human UBQLN1 siRNA respectively was constructed as described previously (9,18). The siRNA sequences used in this experiment as following: UBQLN1 (Gene ID: 29979): 5'-GAAGAAAUCUCUAAACGUUUUUU-3' and 5'-GUACUACUGCGCCAAAUUU-3', non-targeting control siRNA: 5'-UAAGGCUAUGAAGAGAUACAA-3' (11). To overexpress wild-type p53 or silence UBQLN1, the cells were cultured in serum-free medium containing adenovirus (MOI: 50) for 6 h and then incubated in growth medium for another 42 h. KD or overexpression efficiency was determined by Western blot.
Cell viability and proliferation assays
MTT and BrdU assays were used to evaluate cell viability and proliferation, respectively. MTT was performed as described previously (19). Cell proliferation was determined by using a BrdU Cell Proliferation ELISA Kit (colorimetric) (ab126556, Abcam, Cambridge, UK) according to the manufacturer’s protocol.
Apoptosis determination
Apoptosis was induced by incubating the cells in serum-free medium for 72 h and determined by a TUNEL Apoptosis Assay Kit (Roche, Basel, Switzerland) following the manufacturer’s protocol. Apoptotic cells were captured by an Olympus FluoView FV1000 Confocal Microscope. Observation was made at least on 1,000 cells and the apoptotic rate (%) was calculated by dividing the total number of apoptotic cells by the total number of intact cells and multiplying by 100.
Proteasome activity assay
The chymotrypsin-like activity of 20S proteasome core was determined by using synthetic fluorogenic peptide substrate Suc-LLVY-AMC (Boston Biochem, Cambridge, MA, USA) as described previously (20,21). The fluorescence intensity was recorded at an excitation wavelength of 350 nm and emission wavelength of 440 nm by using a fluorescence spectrometer (Perkin Elmer precisely LS55, Billerica, MA, USA).
DHE staining
The real-time formation of intracellular reactive oxygen species (ROS) was measured by dihydroethidium (DHE) staining (22). Briefly, the cells were grown on the coverslips within a 24-well plate at a density of 2×104 cells/well and stained with DHE (at a final concentration of 10 mM) at 37 °C for 30 min in the dark. DHE fluorescence was captured with a fluorescence microscopy (Perkin Elmer precisely LS 55, Billerica, MA, USA).
Western blot
The total proteins were extracted by ice cold lysis buffer (containing 50 mM Hepes, pH7.6, 150 mM NaCl, 1% Triton X-100, 1 mM NaF, 20 mM sodium pyrophosphate, 20 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 10 µg/mL leupeptin, 10 µg/mL aprotinin, 1 µM microcystin-LR, and 1 mM phenylmethylsulfonyl fluoride). The bicinchoninic acid (BCA) assay was used to determine protein concentration. Western blot was performed as described previously (19). The intensity of protein bands was quantified using Bio-Rad Quantity One software.
Statistical analysis
Each experiment was repeated at least 4 times. All data are represented as the mean ± SD. Differences between mean values were examined using the paired Student’s t-test. Chi-square was used to test differences between two or more actual samples. P<0.05 was regarded as statistically significant. The quantification of the relative increase in protein expression and phosphorylation statuses was normalized with control protein expression in each experiment.
Results
UBQLN1 KD promoted the survival and proliferation of A549 cells
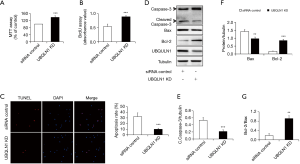
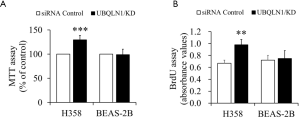
To observe the impacts of UBQLN1 on cell viability and proliferation, UBQLN1-silenced and its control A549 cells were grown in RPMI medium containing 10% FBS for 24 h. As shown in Figure 1A,B, UBQLN1 KD significantly increased cell viability and cell proliferation. The similar observation was made in UBQLN1-KD H358 cells (Figure S1). However, UBQLN1 depletion did not affect cell viability and proliferation in normal non-cancerous BEAS-2B cells (Figure S1). Then, apoptosis was induced by incubating UBQLN1-silenced and its control A549 cells in serum-free RPMI medium for 72 h. TUNEL assay revealed that UBQLN1 KD protected A549 cells against serum-free-induced apoptosis (Figure 1C). Consistent with this finding, UBQLN1-KD cells showed the decreased expressions of cleaved caspase-3 and the increased ratio of Bcl-2/Bax (Figure 1D,E,F,G). These results suggested that UBQLN1 KD resulted in an increase in cell viability and proliferation, and a decrease in cell apoptosis. Moreover, these promotive effects on cell viability and proliferation were cell type dependent.
p53-mediated UBQLN1 action on the survival and proliferation of A549 cells
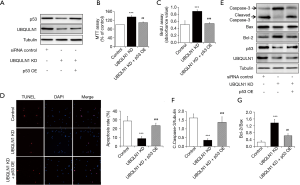
Through scanning the protein which might affect cell survival and proliferation by Western blot (data not shown), we found that UBQLN1 depletion markedly reduced the expression levels of p53 protein, one of the classical type tumor suppressors. To investigate the role of p53 in UBQLN1 regulation on cell survival and proliferation, A549 cells were co-infected with adenovirus/p53 and adenovirus/UBQLN1 siRNA. As shown in Figure 2A, p53 overexpression reversed UBQLN1’s effects on p53 protein levels. Importantly, p53 overexpression significantly mitigated the impacts of UBQLN1 KD on cell viability (Figure 2B), proliferation (Figure 2C), and apoptosis (Figure 2D,E,F,G), suggesting the involvement of p53 in UBQLN1 action on cell survival and proliferation in A549 cells.
Autophagy was responsible for UBQLN1 KD-reduced p53 levels in A549 cells
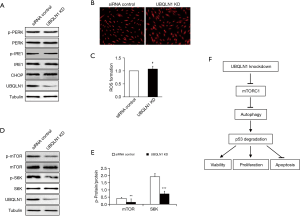
To figure out the mechanism underlying p53 reduction by UBQLN1 silencing, the activities of proteasome and autophagy were investigated in UBQLN1-KD A549 cells. UBQLN1-silenced and its control A549 cells were grown in RPMI medium containing 10% FBS for 24 h. As shown Figure 3A, UBQLN1 KD did not affect proteasome activity. However, UBQLN1-KD cells displayed an increase in LC3-II protein levels (Figure 3B), the ratio of LC3-II to LC3-I (Figure 3C), and the autophagic flux (Figure 3D,E), suggesting that UBQLN1 KD enhanced autophagy activity in A549 cells. When the cells were treated with 20 nM of brefeldin A (BFA), a specific inhibitor of autophagy for 24 h, we found that BFA administration obviously reversed the impacts of UBQLN1 on p53 protein levels (Figure 3F,G). The effects of UBQLN1 depletion on autophagy and p53 protein levels were further confirmed in H358 cells (Figure S2). These results indicated that UBQLN1-KD-enhanced autophagy activity contributed to the reduction of p53 protein levels.

UBQLN1 depletion activated mTOR signaling
Next, we investigated the effects of UBQLN1 KD on ER stress and ROS formation in A549 cells. A549 cells were grown in RPMI medium containing 10% FBS for 24 h. As shown in Figure 4, UBQLN1 KD did not affect ER stress (Figure 4A) and intracellular ROS formation (Figure 4B,C). Interestingly, we found that UBQLN1 KD led to the decreases in the phosphorylation of mTOR and its downstream S6K (Figure 4D,E), suggesting UBQLN1 KD suppressed mTOR signaling pathway.
Discussion
Previous studies have evidenced the promotive effects of UBQLN1 loss on the growth of NSCLC (10,11). Consistent with these studies, we found that UBQLN1 depletion promoted cell viability and proliferation, and suppressed cell apoptosis (Figures 1,S1). In the present study, we also found that UBQLN1 KD reduced p53 protein levels and overexpression of p53 could reversed the impacts of UBQLN1 on cell viability, proliferation, and apoptosis (Figure 2). In addition, loss of UBQLN1 markedly inhibited mTOR phosphorylation (Figure 4) and activated autophagy activity (Figures 3B,C,D,E,S2A,B). Inhibition of autophagy by its specific inhibitor BFA restored UBQLN1 KD-reduced p53 protein levels (Figures 3F,G,S2C,D). Therefore, our results indicated that UBQLN1 silencing suppressed mTOR signaling leading to the activation of autophagy and subsequent reduction of p53 protein, by which UBQLN1 depletion regulated cell viability, proliferation, and apoptosis.
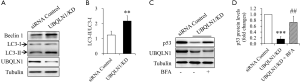
Several mechanisms have been proposed to explain the loss of p53 protein in cancer cells. Generally, p53 is subjected to ubiquitination by a number of E3 ubiquitin ligases and then undergoes a proteasome-dependent degradation (23). Interestingly, the ubiquitinated p53 can function as a signal to regulate some cellular events such as nuclear export as well as transcriptional activation (23). In addition, autophagy and chaperone can also degrade wild type and mutant p53 in cancer cell, although underlying mechanisms are unknown (24-27). In the present study, we found that UBQLN1 depletion did not affect proteasome activity (Figure 3A), which is consistent with previous study (9). Our results also showed that UBQLN1 KD activated autophagy and inhibition of autophagy restored UBQLN1-reduced p53 protein levels (Figures 3B,C,D,E,F,G,S2C,D), suggesting that UBQLN1-induced degradation of p53 was mediated by autophagy. Regardless of the mechanism, autophagy inhibitors are expected to have anticancer activity in part by functioning as p53 activators (28).
There is an interaction between UBQLN1 and autophagy. UBQLN1 can be degraded by autophagy and chaperone-mediated autophagy (CMA) (29). On the other hands, UBQLN1 colocalizes with the autophagosome marker LC3, therefore involving in autophagosome assembly, formation, and maturation (29,30). Deficiency of UBQLN1 decreases, and overexpression of UBQLN1 increases, starvation-induced autophagy (30). The mutants of UBQLN display defective autophagic flux due to suppressed lysosome acidification (31). Recently, the mTOR signaling has been recognized as a key role in UBQLN1 regulation of autophagy. Enhancement of ER stress and ROS formation by UBQLN1 depletion can impair mTOR complex 1 (mTORC1) activity leading to activation of autophagy (31). In the present study, UBQLN1 depletion significantly reduced mTOR phosphorylation (Figure 4C,D) and enhanced autophagy activity (Figure 3B,C,D,E,F,G), but the levels of ER stress markers and ROS formation remained unchanged (Figure 4A,B). These findings indicate that the impacts of UBQLN1 depletion on mTOR or autophagy are independent on ER stress and ROS. The previous study has confirmed that UBQLN1 can directly bind to mTOR (4). In addition, UBQLN1 has been proven to impair mTORC1 activity (31). Hence, the direct regulation of UBQLN1 on mTOR signaling might contribute to its modulation on autophagy activity. More studies are required to reveal the underlying mechanism.
Taken together, we demonstrated that UBQLN1 exerts its impacts on cell viability, proliferation, and apoptosis in A549 cells through impairing mTORC1 activity, promoting autophagy, and degrading p53 (Figure 4F). Thus, UBQLN1 might serve as a useful diagnostic tool for NSCLC and a promising therapy target for treatment of NSCLC. For example, NSCLC is highly expressed some microRNAs like miR-155, which can target UBQLN1 leading to tumorigenesis and progression of NSCLC (10). It should be reasonable to assume that administration with miR-155 inhibitor will increased UBQLN1 levels and suppressed UBQLN1 deficiency-induced promotive effects on cancer growth. More studies are required to confirm this possibility.
Acknowledgments
Funding: This study was supported by Hubei Provincial Natural Science Foundation (No. 2019CFB303).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-1362
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1362
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-1362
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1362). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ko HS, Uehara T, Tsuruma K, et al. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett 2004;566:110-4. [Crossref] [PubMed]
- Lee DY, Brown EJ. Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem 2012;393:441-7. [Crossref] [PubMed]
- Marin I. The ubiquilin gene family: evolutionary patterns and functional insights. BMC Evol Biol 2014;14:63. [Crossref] [PubMed]
- Wu S, Mikhailov A, Kallo-Hosein H, et al. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta 2002;1542:41-56. [Crossref] [PubMed]
- Kurlawala Z, Dunaway R, Shah PP, et al. Regulation of insulin-like growth factor receptors by Ubiquilin1. Biochem J 2017;474:4105-18. [Crossref] [PubMed]
- Adegoke OO, Qiao F, Liu Y, et al. Overexpression of Ubiquilin-1 Alleviates Alzheimer's Disease-Caused Cognitive and Motor Deficits and Reduces Amyloid-beta Accumulation in Mice. J Alzheimers Dis 2017;59:575-90. [Crossref] [PubMed]
- Safren N, Chang L, Dziki KM, et al. Signature changes in ubiquilin expression in the R6/2 mouse model of Huntington's disease. Brain Res 2015;1597:37-46. [Crossref] [PubMed]
- Liu Y, Lu L, Hettinger CL, et al. Ubiquilin-1 protects cells from oxidative stress and ischemic stroke caused tissue injury in mice. J Neurosci 2014;34:2813-21. [Crossref] [PubMed]
- Hu C, Tian Y, Xu H, et al. Inadequate ubiquitination-proteasome coupling contributes to myocardial ischemia-reperfusion injury. J Clin Invest 2018;128:5294-306. [Crossref] [PubMed]
- Yadav S, Singh N, Shah PP, et al. MIR155 Regulation of Ubiquilin1 and Ubiquilin2: Implications in Cellular Protection and Tumorigenesis. Neoplasia 2017;19:321-32. [Crossref] [PubMed]
- Shah PP, Lockwood WW, Saurabh K, et al. Ubiquilin1 represses migration and epithelial-to-mesenchymal transition of human non-small cell lung cancer cells. Oncogene 2015;34:1709-17. [Crossref] [PubMed]
- Bao J, Jiang X, Zhu X, et al. Clinical significance of ubiquilin 1 in gastric cancer. Medicine (Baltimore) 2018;97:e9701. [Crossref] [PubMed]
- Wang Y, Lu J, Zhao X, et al. Prognostic significance of Ubiquilin1 expression in invasive breast cancer. Cancer Biomark 2015;15:635-43. [Crossref] [PubMed]
- Sun Q, Liu T, Yuan Y, et al. MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int J Cancer 2015;136:1003-12. [Crossref] [PubMed]
- Mogi A, Kuwano H. TP53 mutations in nonsmall cell lung cancer. J Biomed Biotechnol 2011;2011:583929. [Crossref] [PubMed]
- Viktorsson K, De Petris L, Lewensohn R. The role of p53 in treatment responses of lung cancer. Biochem Biophys Res Commun 2005;331:868-80. [Crossref] [PubMed]
- Zhang X, Wang Q, Xu Y, et al. lncRNA PCAT19 negatively regulates p53 in non-small cell lung cancer. Oncol Lett 2019;18:6795-800. [Crossref] [PubMed]
- Riccioni T, Cirielli C, Wang X, et al. Adenovirus-mediated wild-type p53 overexpression inhibits endothelial cell differentiation in vitro and angiogenesis in vivo. Gene Ther 1998;5:747-54. [Crossref] [PubMed]
- Yao X, Liu H, Zhang X, et al. Cell Surface GRP78 Accelerated Breast Cancer Cell Proliferation and Migration by Activating STAT3. PLoS One 2015;10:e0125634. [Crossref] [PubMed]
- Li M, Zhang Y, Cao Y, et al. Icariin Ameliorates Palmitate-Induced Insulin Resistance Through Reducing Thioredoxin-Interacting Protein (TXNIP) and Suppressing ER Stress in C2C12 Myotubes. Front Pharmacol 2018;9:1180. [Crossref] [PubMed]
- Ye M, Qiu H, Cao Y, et al. Curcumin Improves Palmitate-Induced Insulin Resistance in Human Umbilical Vein Endothelial Cells by Maintaining Proteostasis in Endoplasmic Reticulum. Front Pharmacol 2017;8:148. [Crossref] [PubMed]
- Jiang P, Zhang D, Qiu H, et al. Tiron ameliorates high glucose-induced cardiac myocyte apoptosis by PKCdelta-dependent inhibition of osteopontin. Clin Exp Pharmacol Physiol 2017;44:760-70. [Crossref] [PubMed]
- Chao CC. Mechanisms of p53 degradation. Clin Chim Acta 2015;438:139-47. [Crossref] [PubMed]
- Guo JY, Karsli-Uzunbas G, Mathew R, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 2013;27:1447-61. [Crossref] [PubMed]
- White E. The role for autophagy in cancer. J Clin Invest 2015;125:42-6. [Crossref] [PubMed]
- Maan M, Pati U. CHIP promotes autophagy-mediated degradation of aggregating mutant p53 in hypoxic conditions. FEBS J 2018;285:3197-214. [Crossref] [PubMed]
- Vakifahmetoglu-Norberg H, Kim M, Xia HG, et al. Chaperone-mediated autophagy degrades mutant p53. Genes Dev 2013;27:1718-30. [Crossref] [PubMed]
- White E. Autophagy and p53. Cold Spring Harb Perspect Med 2016;6:a026120. [Crossref] [PubMed]
- Rothenberg C, Srinivasan D, Mah L, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet 2010;19:3219-32. [Crossref] [PubMed]
- N'Diaye EN, Kajihara KK, Hsieh I, et al. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 2009;10:173-9. [Crossref] [PubMed]
- Senturk M, Lin G, Zuo Z, et al. Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat Cell Biol 2019;21:384-96. [Crossref] [PubMed]


