Genomic alterations of whole exome sequencing in esophageal squamous cell carcinoma before and after radiotherapy
Introduction
Esophageal carcinoma (EC) is among the most fatal malignancies in the world, with a 5-year survival rate of less than 10% (1). EC’s are classified as esophageal adenocarcinomas (EAC) and esophageal squamous cell carcinomas (ESCC), according to the histologic subtype of tumor cells (2). ESCC is especially prominent in China, where it accounts for approximately 88.8% of all esophageal cancer cases (3). Surgical resection is the most common treatment for respectable ESCC (4), and postoperative adjuvant chemotherapy, radiotherapy, and chemoradiotherapy are also routinely practiced (5). Although radiotherapy has significantly improved the median survival period the recurrence rate of ESCC remains high (6,7). Therefore a better understanding of tumoral radiosensitivity is urgent needed to develop strategies and further personalize radiation treatments. Thus, we performed whole-exome sequencing (WES) on paired tumors collected before and after radiotherapy from 11 patients with ESCC. Comprehensive analysis the somatic mutations, the driver genes mutations, the copy number variations (CNVs), the mutational signatures, the tumor’s clonal composition, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway between pre- and post-radiotherapy samples in our cohort.
In recent years, several genes have been found to play an important role in radiosensitivity. Genetic alteration of cluster of differentiation 59 (CD59) expression modulated the radiosensitivity of esophageal cancer cells to ionizing radiation (8). The downregulation of the high mobility group box 1 (HMGB1) gene could effectively inhibit proliferation, increase radiosensitivity, impair deoxyribonucleic acid (DNA) damage-repair abilities, reduce autophagy, and increase apoptosis rates in ESCC cells after irradiation (9).
Most of the previous studies exploring the molecular mechanisms underlying radiotherapy resistance have been limited to one or several specific biomarkers by in vitro experiments, and none are currently used in the prediction of radiosensitivity in clinical practice (10). The genomic landscape of ESCC has been characterized (11,12), revealing that pathogenesis is caused by a large number of genomic alterations through single-nucleotide variations and/or copy number losses. However, the changes of mutational processes in ESCC across the whole-genome due to radiotherapy remain unknown. Though a similar study identified ESCC mutational heterogeneity by Single-cell whole-exome sequencing (scWES), scWES inevitably encounter much more experimental error and analysis error than WES. In the current study, we performed WES of 11 matched pre- and post-radiotherapy ESCC tumors from Chinese individuals. We examined the single-nucleotide variants (SNVs) spectrum, mutational/trinucleotide context, and copy-number aberrations in this cohort. We aimed to comprehensively evaluate the impact of radiotherapy on the exome landscape and possibly identify potential actionable targets to increase sensitivity and avoid unnecessary toxicity of ESCC radiotherapy.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2450).
Methods
Patients
ESCC tumor samples and adjacent, histologically normal tissue samples from 11 patients before and after radiotherapy were collected in Shandong Cancer Hospital from 2015 to 2017. We acquired the written informed consent from these patients. Three of them received surgery, and 10 of them received chemotherapy, and only one patient received Nimotuzumab. All the patients received radiotherapy. All samples were subjected to hematoxylin and eosin staining and histopathological examination to separate the normal cells from the tumor cells (Figure S1). The isolated normal cells were used as matched normal controls. Only one normal control was selected for each patient.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by Shandong Cancer Hospital and Institute (Approval ID: SDTHEC201604003).
DNA extraction and Next-generation sequencing
The genomic DNA were extracted from the archived formalin-fixed paraffin-embedded (FFPE) samples of 11 pairs ESCC tumor tissue and matched normal tissue according to the manufacture’s instruction (K182001, Invitrogen™ PureLink™ Genomic DNA Mini Kit, China). Then, qualified DNA were sonicated to an average size of 200 base pairs (bp) (range, 100–500 bp) (13). The targeted DNA fragments were captured pull-down. Exon-wide libraries were created using the xGen® Exome Research Panel (Integrated DNA Technologies, Inc., Illinois, USA) and TruePrep DNA Library Prep Kit V2 for Illumina (#TD501, Vazyme, Nanjing, China), and paired-end sequence data was generated using Illumina HiSeq machines.
Sequence alignment and variant calling
The sequence data was aligned to the human reference genome [National Centre for Biotechnology Information (NCBI) build 37] using Burrows-Wheeler Aligner (BWA) (14). Sambamba was employed to sort and remove polymerase chain reaction (PCR) duplicates (15). Base quality score recalibration (BQSR) was performed using the genome analysis toolkit (GATK) 4.1.2 (16). The panel of normal samples was first created with Mutect2 (16), and subsequently used for mutation analysis of paired and uncontrolled samples. The sequencing data of matched normal samples were utilized to exclude both germline mutations and sequencing artifacts during detection of somatic mutations, which could increase the true positive rates of the calling. Somatic mutation calling was performed using Strelka2 (17), Mutect2, and VarDict (18). Somatic mutations present in at least two of these three aforementioned software programs were selected as high-confidence mutations. The single-sample mode of Mutect2 and VarDict was used for samples without normal tissues (library construction of the three normal samples failed due to the low concentration of DNA), and the intersection was then calculated. The variant data was annotated using ANNOVAR (19). Somatic mutations were converted to mutation annotation format (MAF) and visualized using R package maftools (20). Gene mutation rate was calculated by the number of tissues with mutation gene divided as the total number of all tissues. Driver gene fold enrichment was analyzed as previously described (20). Copy number variations (CNVs) were called using CNVkit (21) with the binary alignment maps (BAMs) after BQSR and germline mutations from VarDict. CNVs were exported to SEG format, and GISTIC2 (22) was used to identify regions that were significantly amplified or deleted across samples. Mutational signatures were analyzed using Mutational Patterns (23).
Phylogenetic analysis of tumor samples
The reads supporting alternate and reference bases of each of the SNVs from the paired tumor samples were calculated using GATK’s module CollectAllelicCounts. The variant allele frequencies of each of the SNVs and the allelic-specific somatic copy numbers from CNVkit were subsequently utilized as input to the SciClone clustering algorithm (24). Regions with somatic copy number alternations and loss of heterozygosity (LOH) were excluded. Phylogenetic trees were constructed using R package ClonEvol (https://www.ncbi.nlm.nih.gov/pubmed/28950321).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
To understand the biological mechanisms of radiotherapy in ESCC, KEGG pathway analyses were performed using ClusterProfiler, which is an R package for functional classification and enrichment of gene clusters using hypergeometric distribution (25).
Statistical analysis
The SPSS Statistics 22.0 package, and ggpubr package (26) in R (27) were employed to correlate clinical and biological variables using Fisher’s exact test or a non-parametric test when necessary. The progression-free survival (PFS) and overall survival (OS) were calculated using the Kaplan-Meier method, and differences in the variables were compared using the log-rank test. Data is shown as the mean ± standard deviation (SD), and a two-tailed P value <0.05 was considered statistically significant.
Results
Patient characteristics
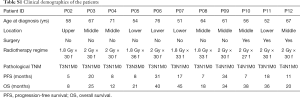
For this study, we collected 22 formalin-fixed paraffin-embedded (FFPE) tumor samples from 11 EC patients before and after radiotherapy. Their baseline characteristics are summarized in Table S1. The most common tumor locations were middle thoracic (54.5%, n=6). All patients were male, and the median age was 61 years (range between 51–76). The total radiation dose ranged from 54–64.8 Gy, and the radiation schedule was similar with an arrangement of 1.8–2 Gy/fraction per day, 5 times/week.
Full table
The mutational landscape of ESCC samples pre- and post-radiotherapy
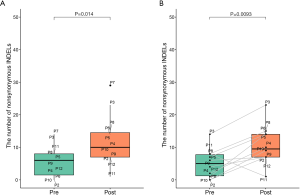
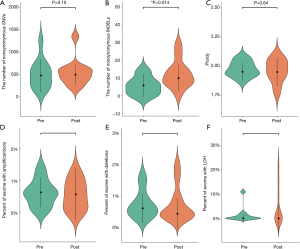
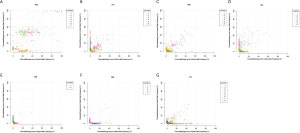
We performed WES on pre- and post-radiotherapy paired samples from 11 patients to identify somatic alterations in ESCC. The mean coverage depth for the tumor and normal samples were 196× and 126×, respectively. The median number of genes with non-synonymous mutations in the pre- and post-radiotherapy samples was 474 and 507, respectively. The average tumor mutation burden (TMB) was similar between the pre-radiotherapy group (12.5 mutations/Mb) and the post-radiotherapy group (13.3 mutations/Mb) (Wilcoxon rank-sum test, P=0.102) (Figure S2A). There were no differences in the number of SNVs, the average ploidy, the proportions of amplifications, deletions, and LOH regions between the two groups (Wilcoxon rank-sum test, P>0.05) (Figure 1). However, more insertions/deletions (indels) were observed in post-radiotherapy samples compared to pre-radiotherapy samples (P=0.014). Among the patients, the indels of patient 7 (P07) varied the most (Figure S2B), and the TMB of P07 was considerably higher after radiotherapy (Figure S2A), implying that this particular patient had significant genomic variations due to radiation. P07 was considered an outlier by box plot (Figure S3A). After removing P07, the total number of indels still increased markedly after radiotherapy (P=0.0093) (Figure S3B).
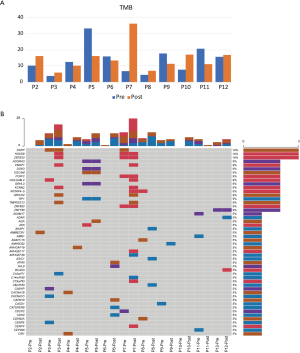
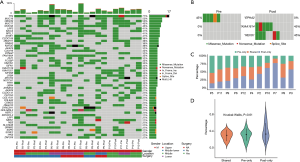
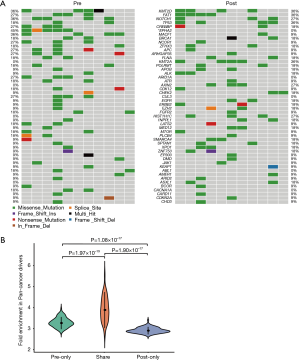
We identified several of the most frequently mutated genes including Tenascin-N (TNN), Mucin 16 (MUC16) and Obscurin (OBSCN), in addition to widespread heterogeneous alterations (Figure 2A, http://fp.amegroups.cn/cms/3a54761e061a9714e70c298177e8da76/jtd-20-2450-1.xlsx). Compared with the pre-radiotherapy samples, the mutation rate of Ephrin-A2 (EPHA2) was significantly reduced (Fisher’s exact test, P=0.035), while the mutation rates of KIAA1614 and RNA Exonuclease Homolog 1 (REXO1) were significantly increased (Fisher’s exact test, two P values were 0.035) (Figure 2B). We also found 30.4% (7.9–52.1%) of SNVs were pre-radiotherapy-specific, 39.6% (15.7–78.2%) of SNVs were postradiotherapy- samples (Figure 2C). Although the number of SNVs showed no differences among the three groups (Kruskal-Wallis, P=0.61) (Figure 2D) .We assessed the mutation frequency of driver genes to compare the overall mutational landscape of the pre- and post-radiotherapy samples in ESCC (28). The result showed that the mutation rates of driver genes were similar in the two groups, except for EPHA2 (Figure 3A). The pan-cancer driver genes were enriched in the shared group (29) (Figure 3B). This suggested that the driver gene mutation occurs before radiotherapy, while only a few additional driver genes are altered during or after radiotherapy.
Copy number variations
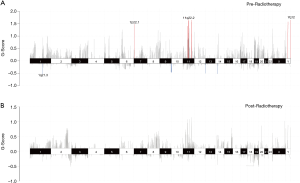
Genomic CNVs were analyzed to further explore radiotherapy-related molecular events. CNVs amplifications were most commonly detected with chromosome regions 7q22.1, 11q22.2, and Yq12, and the most frequently deleted chromosome region in pre-radiotherapy samples was 1q21.3 (Figure 4). No significant CNVs were detected in post-radiotherapy samples. However, chromosome regions with CNVs did not harbor any driver genes, indicating that CNVs occurred in passenger genes.
Phylogenetic analysis
The clonal evolution analysis identified two modes of clonal composition among nine patients. One mode showed similar clonal compositions between pre- and post-radiotherapy, typically represented by P05 (Figure 5A,B). On the basis of mutation clustering results, we inferred the identity of five clones with distinct sets of mutations (clusters) in the pre-radiotherapy tumor. Four clusters were found in both pre- and post-radiotherapy samples. One cluster, which presented only in pre-radiotherapy samples, was eliminated under the evolutionary force after radiotherapy. Most shared clusters implied that these tumor cells were not sensitive to radiation. This pattern was also identified in four other pre- and post-radiotherapy pairs (P03, P04, P09, and P12) (Figure S4A,B,C,D).
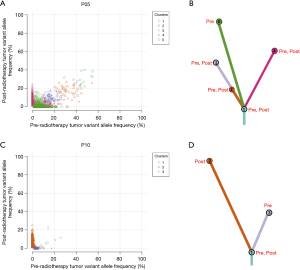
The other mode showed completely different clonal compositions between pre- and post-radiotherapy, typically represented by P10 (Figure 5C,D). One mutation cluster was identified in the pre-radiotherapy tumor, and the other was found in the post-radiotherapy tumor. Only one small cluster (cluster 1) containing five mutations was shared in both samples. It is likely that cluster 2 mutations have evolved to become the dominant clone by gaining radiotherapy-specific mutations, while cluster 3 mutations almost disappeared after radiotherapy. We suspect that cluster 2 mutations in this clone provided a strong selective advantage after radiotherapy. Clonal evolution analyses revealed that pre- and post-radiotherapy tumors exhibited entirely distinct evolutionary processes. This pattern was also observed in three other pre- and post-radiotherapy pairs (P02, P06, and P11) (Figure 4E,F,G).
Mutational spectrum and mutational signatures during radiotherapy
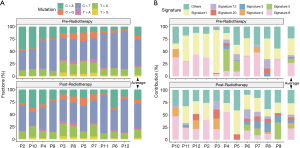
The six subtypes of base substitutions (C > A, C > G, C > T, T > A, T > C, and T > G) were unevenly presented in the SNVs (Figure 6A). C > T was the most common substitution in ESCC (44.4% in pre-radiotherapy and 44.8% in post-radiotherapy), consistent with the previous report (30). Signatures 1 and 6 were identified as dominant in both pre- and post-radiotherapy samples (Figure 6B). Some mutational signatures differed in the pre- and post-radiotherapy of the same patient, such as patient 3 (P03), with dominant signature 1 (in pre-radiotherapy) and dominant signatures 4 and 6 (in post-radiotherapy). However, the comparison of average mutational spectra and signatures for the 11 pairs of pre- and post-radiotherapy samples did not exhibit any clear differences (Wilcoxon rank-sum test, P>0.05).
KEGG pathway enrichment analysis
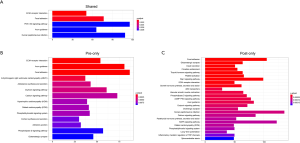
We divided SNVs into shared SNVs, pre-radiotherapy-specific SNVs, and post-radiotherapy-specific SNVs to explore the functional impact of the mutations. We subsequently analyzed the KEGG pathway enrichment of the altered genes with these SNVs (Figure S5). We found that tumor-related pathways, such as the extracellular matrix (ECM)-receptor interaction and the Focal adhesion, existed in all three groups, and the phosphatidylinositol-3-kinase -protein kinase B (PI3K-Akt) signaling pathway existed in the shared group. Adherens junction only existed in the pre-radiotherapy-specific group, and the Ras-proximate-1 (Rap1), cyclic guanosine monophosphate-protein kinase G (cGMP-PKG), and the mitogen-activating protein kinase (MAPK) signaling pathways only existed in the post-radiotherapy-specific group.
Survival analysis by genotype
Survival analysis was conducted for all gene mutations to identify the independent prediction factor for outcomes. In pre-radiotherapy samples, the mutations of 240 genes were associated with shorter PFS, and mutations in 235 genes were associated with shorter OS (Log-Rank test, P<0.05) (http://fp.amegroups.cn/cms/85cb0833c11f8c36bdd443abb0fbe39e/jtd-20-2450-2.pdf and http://fp.amegroups.cn/cms/65f3050e0aee835d1fd060c8218c2d90/jtd-20-2450-3.pdf). Several driver genes were found in both tables, including asparaginyl-TRNA synthetase (NARS), SIN3 homolog A (SIN3A), and tuberous sclerosis 1 (TSC1).
Discussion
ESCC is among the most fatal malignancies worldwide, with increasing incidences and poor outcomes. Radiotherapy is a well-established treatment for various cancer types, including ESCC. In this study, we have systematically analyzed whole-exome copy number, SNVs, mutation spectrum, and mutational signatures by WES in order to establish a more widespread genomic landscape of radiotherapy in ESCC.
We found that the mutation rate of EPHA2 was significantly reduced in the post-radiotherapy samples, suggesting that the loss of EPHA2 mutations may provide a selective advantage to ESCC cells under radiotherapy stress. EPHA2, a receptor tyrosine kinase, is implicated in tumor progression and is considered a potential therapeutic target in metastatic cancer (31). The deletion of EPHA2 was found to reverse T-cell exclusion and sensitize tumors to immunotherapy (32). Also, our study found that TMB was high, indicating that it is rational to apply immune checkpoint inhibitors in ESCC patients since they might be susceptible to immunotherapy (33). This is especially true for patients with deletion of EPHA2. Though, further experiments regarding this problem are needed.
The most common mutated driver genes in our study included Lysine K-specific methyltransferase 2D (KMT2D), FAT atypical cadherin 1 (FAT1), Notch homolog 1 (NOTCH1), and tumor protein 53 (TP53), among others (Figure 3A and http://fp.amegroups.cn/cms/3a54761e061a9714e70c298177e8da76/jtd-20-2450-1.xlsx). High-frequency mutations in these genes have also been detected in previous studies (30,34,35). However, the frequency of TP53 mutation in our study (27.3% in all samples) was significantly lower than that in previous studies (59.7–93%). This may be due to our relatively strict screening criteria; somatic mutations in at least two of the three software programs (Strelka2, Mutect2, and VarDict) were selected as high-confidence mutations. Some low-frequency mutations may not appear in our results. On the other hand, the sample size in our cohort was small, which may have also led to the discrepancy.
The total number of indels was greater in the post-radiotherapy group compared to the pre-radiotherapy group, and was typically seen in two patients (P06 and P07). However, differences in the number of indels and CNVs were not prevalent in the driver gene. This may indicate new mechanisms underlying the interplay between these genomic variants and the tumor, which cannot be captured by the known driver gene. Additional experiments are required to further elucidate this process.
We identified that signatures 1 and 6 were the most common mutational signatures in our ESCC samples. Signature 1 exhibits strong positive correlations with age in most cancers, including ESCC (36). Signature 6, which contributed a large number of substations and small indels at repeat sequence, was associated with defective DNA mismatch repair (MMR) and was found in “microsatellite instability” cancers, including ESCC (36). We also detected mutation in DNA MMR genes including MutL Homolog 1 (MLH1), PMS1 Homolog 2 (PMS2), and MutS Homolog 6 (MSH6) (http://fp.amegroups.cn/cms/3a54761e061a9714e70c298177e8da76/jtd-20-2450-1.xlsx), which might contribute to signature 6. These results demonstrated the importance of DNA MMR deficiency in ESCC. However, the comparison of mutational signatures for pre- and post-radiotherapy did not exhibit any clear difference. KEGG pathway enrichment analysis showed that the Rap1, cGMP-PKG, and the MAPK signaling pathways only existed in the post-radiotherapy-specific group. These pathways are potentially involved in radiotherapy resistance.
From the clonal analysis of our study, we inferred that radiotherapy had two modes of effect on the clonal architecture of ESCC. First, almost all clones were present in both pre- and post-radiotherapy samples, as was observed in five patients (P03, P04, P05, P09, and P12). Second, only a small cluster was shared by both the pre- and post-radiotherapy samples in 4 patients (P02, P06, P10, and P11), signifying that the new clone survived and evolved to become the dominant clone after radiotherapy. For tumors harboring both radiosensitive and non-radiosensitive clones, the previously predominant radiosensitive clones perished and were replaced by non-radiosensitive clones after radiotherapy.
Also, we observed that the majority of ESCC genomes remained rather stable following radiotherapy. Research regarding the genomic effects in EAC of chemotherapy revealed no significant differences in the overall mutation rate, mutation signatures, specific recurrent point mutations, or copy-number events with respect to chemotherapy status (37). Therefore, it is possible that a considerable number of EC patients are not sensitive to radiotherapy or chemotherapy, owing to intensive intratumoral heterogeneity. Poor survival would support the findings that this cancer is resistant to radiotherapy and chemotherapy with remarkable consistency in the genome of the primary tumor over time. It is worth noting that any of these conclusions are subject to possible change in the future should additional findings become apparent through analyses of radiotherapy of whole genome sequences in ESCC. This is especially true considering that radiation-induced genomic alterations may occur in the intron region, and not in the exon region. On the other hand, gene expression is determined not only by genetic variation in the promoter and other regulatory elements but also by epigenetic modification of transcriptional activity and post-translational modulation (38). Radiation can induce genetic and epigenetic changes in cancer cells that confer stem-like properties and radioresistance in these cells (39). In addition, the methylation status may influence cellular radiation response, while WES would not reveal epigenetic changes in gene expression. Furthermore, females are more sensitive to radiotherapy than males (40), while esophageal cancer is more common in men than women (41). Considering that all of the cases in our study cohort were male, a bias potentially existed, which to some extent also affected the results.
We identified that mutations in 240 genes were associated with shorter PFS, and mutations in 235 genes were associated with shorter OS (log-rank test). However, we also noted that all these genes were mutated in two patients who had a very short PFS and OS, and the reliability of the survival analysis was questionable due to the limited number of cases.
In conclusion, we conducted the genomic profile of ESCC before and after radiotherapy and identified novel radiotherapy-related mutations, which provided potential biomarkers for prognosis and targeted therapeutics. In the future, we plan to bring into more technology advertisement and larger cohort prospective studies to revel the molecular mechanisms and genetic alterations in ESCC radioresistance.
Acknowledgments
The authors thank Shanghai Tongshu Biotechnology Co., Ltd. for their technical support.
Funding: This study was partially funded by the Natural Science Foundation of China (NSFC81872475, NSFC81372413), the Shandong Key Research and Development Plan (2017CXGC1209 and 2017GSF18164), the Outstanding Youth Natural Science Foundation of Shandong Province (JQ201423), the Jinan Clinical Medicine Science and Technology Innovation Plan (201704095), and the National Key Research and Development Program of China (2016YFC0904700).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2450
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2450
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2450). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by Shandong Cancer Hospital and Institute (Approval ID: SDTHEC201604003). Informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg 2018;41:210-5. [Crossref] [PubMed]
- Chen D, Fan N, Mo J, et al. Multiple primary malignancies for squamous cell carcinoma and adenocarcinoma of the esophagus. J Thorac Dis 2019;11:3292-301. [Crossref] [PubMed]
- Zhang H, Yang Y, Shang Q, et al. Predictive value of preoperative weight loss on survival of elderly patients undergoing surgery for esophageal squamous cell carcinoma. Transl Cancer Res 2019;8:2752-8. [Crossref]
- van der Horst S, de Maat MFG, van der Sluis PC, et al. Extended thoracic lymph node dissection in robotic-assisted minimal invasive esophagectomy (RAMIE) for patients with superior mediastinal lymph node metastasis. Ann Cardiothorac Surg 2019;8:218-25. [Crossref] [PubMed]
- Ohashi S, Miyamoto S, Kikuchi O, et al. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology 2015;149:1700-15. [Crossref] [PubMed]
- Berger B, Belka C. Evidence-based radiation oncology: oesophagus. Radiother Oncol 2009;92:276-90. [Crossref] [PubMed]
- Cui H, Zhang S, Zhou H, et al. Direct Downregulation of B-Cell Translocation Gene 3 by microRNA-93 Is Required for Desensitizing Esophageal Cancer to Radiotherapy. Dig Dis Sci 2017;62:1995-2003. [Crossref] [PubMed]
- Zhou Y, Chu L, Wang Q, et al. CD59 is a potential biomarker of esophageal squamous cell carcinoma radioresistance by affecting DNA repair. Cell Death Dis 2018;9:887. [Crossref] [PubMed]
- Di X, He G, Chen H, et al. High-mobility group box 1 protein modulated proliferation and radioresistance in esophageal squamous cell carcinoma. J Gastroenterol Hepatol 2019;34:728-35. [Crossref] [PubMed]
- Liauw SL, Connell PP, Weichselbaum RR. New paradigms and future challenges in radiation oncology: an update of biological targets and technology. Sci Transl Med 2013;5:173sr2. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91-5. [Crossref] [PubMed]
- Chen XX, Zhong Q, Liu Y, et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat Commun 2017;8:524. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Tarasov A, Vilella AJ, Cuppen E, et al. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032-4. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591-4. [Crossref] [PubMed]
- Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44:e108. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Hu Z, Ding J, Ma Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 2019;51:1113-22. [Crossref] [PubMed]
- Talevich E, Shain AH, Botton T, et al. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12:e1004873. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Blokzijl F, Janssen R, van Boxtel R, et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [Crossref] [PubMed]
- Miller CA, White BS, Dees ND, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014;10:e1003665. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Kassambara A. ggpubr: 'ggplot2' Based Publication Ready Plots. 2018. Available online: http://www.sthda.com/english/rpkgs/ggpubr.
- Team RC. R: A Language and Environment for Statistical Computing. 2018. Available online: https://www.R-project.org/
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;173:371-85.e18. [Crossref] [PubMed]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;174:1034-5. [Crossref] [PubMed]
- Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015;96:597-611. [Crossref] [PubMed]
- Martini G, Cardone C, Vitiello PP, et al. EPHA2 Is a Predictive Biomarker of Resistance and a Potential Therapeutic Target for Improving Antiepidermal Growth Factor Receptor Therapy in Colorectal Cancer. Mol Cancer Ther 2019;18:845-55. [Crossref] [PubMed]
- Markosyan N, Li J, Sun YH, et al. Tumor cell-intrinsic EPHA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2). J Clin Invest 2019;129:3594-609. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014;46:467-73. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Noorani A, Bornschein J, Lynch AG, et al. A comparative analysis of whole genome sequencing of esophageal adenocarcinoma pre- and post-chemotherapy. Genome Res 2017;27:902-12. [Crossref] [PubMed]
- Herskind C, Talbot CJ, Kerns SL, et al. Radiogenomics: A systems biology approach to understanding genetic risk factors for radiotherapy toxicity? Cancer Lett 2016;382:95-109. [Crossref] [PubMed]
- Peitzsch C, Cojoc M, Hein L, et al. An Epigenetic Reprogramming Strategy to Resensitize Radioresistant Prostate Cancer Cells. Cancer Res 2016;76:2637-51. [Crossref] [PubMed]
- Gasinska A, Darasz Z, Adamczyk A, et al. Gender-related prognostic significance of clinical and biological tumor features in rectal cancer patients receiving short-course preoperative radiotherapy. Rep Pract Oncol Radiother 2017;22:368-77. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
(English Language Editor: A. Kassem)