Expression of paired basic amino acid-cleaving enzyme 4 (PACE4) correlated with prognosis in non-small cell lung cancer (NSCLC) patients
Introduction
Primary lung cancer is the leading cause of cancer mortality worldwide. Although optimal treatment of surgical resection for early-stage, non-small cell lung cancer (NSCLC), 5-year survival rates for surgically resectable NSCLC are still unsatisfactory and range from 19% for stage IIIA to 63% for stage IA (1). Therefore, the identification of predictive and/or prognostic markers is important to stratify patients with resected NSCLC and select high-risk patients who should receive aggressive adjuvant chemotherapy.
The proprotein convertases (PCs) are a family of Ca2+-dependent serine proteases involved in processing and activation of several types of substrates (2). It comprises nine members: PC1/3, PC2, furin, PC4, PC5/6, PACE4, PC7, SKI-1/S1P, and PCSK9 (3,4). There is growing evidence for the involvement of PCs in various cancer types, including roles in each of the biologic capabilities acquired during the multistep development of human tumors (5). These biologic capabilities include proliferative signaling, evading growth suppressors, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis (6-10). Overexpression of PCs has been documented in a variety of neoplasms, including those from skin, ovary, nervous system, colon, and breast cancer (11-15). Monitoring the total PC activity in uterine lavage may provide a rapid and non-invasive method for the diagnosis of endometrial cancer in postmenopausal women, and show a complex association between individual PCs and endometrial cancer (16).
Paired basic amino acid-cleaving enzyme 4 (PACE4), also call PCSK6, proprotein convertase subtilisin/kexin type 6. The tissue distribution of PACE4 was widespread, with comparatively higher levels in the liver. The close proximity of the fur and PACE4 genes on chromosome 15 suggests that these genes probably evolved from a common ancestor by gene duplication (17). The human PACE4 gene spans at least 250 kb and is distributed over 25 exons that range in size from 39 to 1,422 base pairs. These results suggest that PACE4 is not a constitutive gene product and its expression is regulated by various transcription factors (18). PACE4 overexpression resulted in enhanced susceptibility to carcinogenesis and tumor progression pointing to a new target for blocking tumor cell invasiveness (10). Knockdown of PACE4 in human breast cancer MDA-MB-231 cells showed significantly reduced proliferation, migration and invasion rates (19). It also confirm the unique role of the proprotein convertase PACE4 in prostate cancer cell proliferation, it has a distinct role in maintaining proliferation and tumor progression in prostate cancer (20,21). Up-regulation of PACE4 expression is associated with SCC conversion to SPCC (22), and participates in bone formation at least in osteoblast differentiation, induce osteoblast differentiation by estrogen receptor-mediated stimuli (23). PACE4 expression was correlated with sustainment of SKOV3 cell proliferation in vitro and in vivo (24). But how PACE4 regulated tumor’s proliferation and invasive? PACE4 expression is specifically unregulated by PDGF-BB in differentiated megakaryoblasts (25). It plays an important role in myogenic differentiation through its association with the IGF-II pathway (26), and may regulate the rate of hypertrophic conversion of ATDC5 cells through activation of proBMP6 (27). There was evidence that CMK-dependent in vivo inhibition of PACE4 activity results in decreased epidermal basal keratinocyte proliferation, tumor development, and metastasis (28).
Our objectives of this study were (I) to immunohistochemically examine PACE4 expression in tumor samples of 172 NSCLC patients, (II) to evaluate the relationships between PACE4 expression and the clinicopathological parameters of NSCLC, and (III) to estimate the prognostic impact of PACE4 on survival in patients with resected NSCLC patients.
Patients and methods
Patients and tissue specimens
A total of 213 paraffin-embedded archived lung cancer samples which were histologically and clinically diagnosed from the Department of Pathology, The First Affiliated Hospital of Guangzhou Medical University, China, between January 2006 and May 2009. The cases selected were based on a distinctive pathologic diagnosis of NSCLC in patients who underwent primary and curative resection, the availability of resected tissue, and follow-up data. None of them had received radiation therapy or chemotherapy before surgery. Prior to the use of these clinical materials for investigation, informed consent from patients and approval from the Institute Research Ethics Committee were obtained. Patients whose cause of death remained unknown were excluded from our study. Of the informative 172 lung cancer tissues, 15 matched adjacent noncancerous tissues were used as controls. The NSCLC cases included 127 (73.8%) men and 45 (26.2%) women, with a mean age of 58.1 years. The average follow-up time was 46 months (median, 36 months; range, 1-111 months). Clinicopathological characteristics for these patients, including age, sex, smoking, lymph node metastasis, tumor size and pathological classification, clinical stage, death, and follow-up time, are detailed in Table 1. The lung adenocarcinomas were reclassified by using the newest guideline (29). Tumor stage was defined according to the American Joint Committee on Cancer/International Union against Cancer tumor-node-metastasis (TNM) classification system (30).
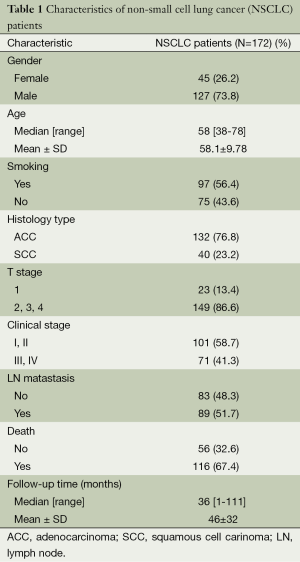
Full table
Two senior pathologists (Q.N.W. and G.Q.L.) reviewed the slides to determine and mark out representative tumor areas. Duplicates of 1-mm-diameter cylinders were punched from representative tumor areas and form the TMA block, which contained 213 lung cancers and 20 adjacent nonmalignant lung tissues (Beecher Instruments, Silver Spring, MD, USA).
Cell lines and cell cultures
The NSCLC lines (A549, SPC-A-1, H1650, H460, H520, and H1975) and human bronchial epithelial cells (16HBE) cell line were purchased from American Type Culture Collection (ATCC, Manassas, USA). They were cultured in RPMI-1640 medium (Gibco, USA) with 10% fetal bovine serum (Gibco, USA). All cell lines were grown in a humidified incubator 37 °C with 5% CO2.
RNA extraction and quantitative RT-PCR analysis
Total RNA NSCLC lines (A549, SPC-A-1, H1650, H460, H520, H1975) and 16HBE cell line using TRIzol reagent (Dongsheng biotech, China). After reverse transcription of the total RNA, the cDNA was then used as templates for detection.
PACE4 expression by quantitative real-time PCR (qRT-PCR) with the SYBR Green I chemistry. Primers of PACE4 were used for the PCR reaction with forward 5'-CTATGGATTTGGTTTGGTGGAC-3'; reverse 5'-AGGCTCCATTCTTTCAACTTCC-3'; GAPDH forward 5'-CTGCACCACCAACTGCTTAG-3'; GAPDH reverse 5'-AGGTCCACCACTGACACGTT-3'. Threshold cycles (Ct) for GAPDH (reference) and PACE4 (sample) were determined in triplicates (shown as arithmetical mean). The quantity of PACE4 in each NSCLC cell line relative to the average expression in 16HBE cells, was calculated using the relative expression levels of PACE4 were given by 2−△△CT, where △Ct = Ct (unknown) − Ct (internal control) (11).
Immunofluorescence assay
Collect the NSCLC lines (A549, SPC-A-1, H1650, H460, H520, and H1975) and 16HBE cell line, which were harvested and washed thrice with PBS, suspended in PBS and centrifuged on the slides. Slides were fixed with 95% alcohols for 30 min, permeabilized. After 1 hour blocking in PBS +0.1% Tween plus 1% bovine serum albumin, cells were incubated with primary antibodies of PACE4 (1:100 dilution, Rabbit mAb, Abcam) at 37 °C 1 hour, then with secondary antibody 30 min at 37 °C. After counterstaining with DAPI (1 µg/mL) for 10 min, slides were observed and photographed with fluorescence microscopy.
Immunohistochemical staining
Commercially available antibody against PACE4 (1:100 diluton, Rabbit mAb, Abcam) was used as the primary antibody. Formalin-fixed, paraffin-embedded NSCLC samples were cut into 4-µm thick sequential sections and baked for 3 hours at 58 °C. Then they were deparaffined in xylene and rehydrated in graded alcohols, treated with 3% hydrogen peroxide for 10 min to quench the endogenous peroxidase activity. Antigen retrieved by pressure cooking for 3 min in EDTA buffer (pH 8.0), then the slides were allowed to cool at room temperature. Next, they were preincubated with 10% normal goat serum at room temperature for 30 min to reduce nonspecific reaction. Subsequently, the slides were incubated with mouse monoclonal anti-PACE4 for 2 hours at room temperature. After washing with phosphate-buffered saline, biotinylated secondary antibody was applied for 15 min at 37 °C. Then the sections were incubated with streptavidin–horseradish peroxidase complex and developed with Diaminobenzidine (DAB). Finally, the sections were counterstained with Mayer hematoxylin, dehydrated, and mounted. For negative controls, the primary antibody was replaced by normal rabbit serum. Known immunostaining positive slides were used as positive controls.
Immunohistochemical evaluation
Two investigators (Q.N.W. and G.Q.L.) separately evaluated all the specimens without knowledge of the clinical features of the cases. Variant cases were reviewed and discussed until a consensus was obtained. Five areas were selected at random and scored. The percentage of tumor cells with positive staining of PACE4 was determined at high magnification (×200). For PACE4, cytoplasmic immunostaining in tumor cells was considered to be positive. Tissue was scored (H score) based on the total percentage of positive cells and the intensity of the staining (1+, 2+, or 3+), where H = (%1+ ×1) + (%2+ ×2) + (%3+ ×3). The sample was considered negative if H=0 and positive if H was more than 0 (31).
Statistical analysis
All statistical analyses were performed using the SPSS 16.0 statistical software package. Because of the non-normal distribution of protein expression, statistical evaluation was performed using nonparametric tests. The correlation between PACE4 expression and clinicopathological status of NSCLC patients was assessed by chi-square test. Survival curves for both PACE4 positive-expression and PACE4 negative-expression patients were plotted using the Kaplan-Meier analysis and log-rank test. Univariate and multivariate regression analysis were performed with the Cox proportional hazards regression model to determine the effect of particular prognostic factors on survival. Further analyses of survival curves were computed based on stratifying histology type and clinical stages. Survival time was calculated from the date of lung cancer diagnosis to the date of death for any cause. In all cases, differences with P<0.05 were considered statistically significant.
Results
High expression of PACE4 in NSCLC tissues
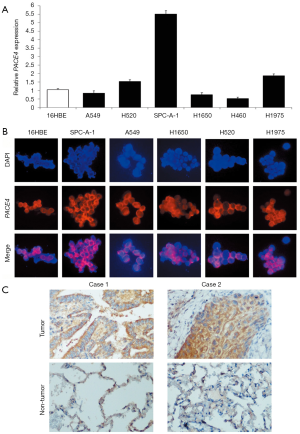
To examine the expression pattern of PACE4 in NSCLC tissues and cells, RT-PCR and immunofluorescence analysis showed that PACE4 expression was higher in NSCLC lines (SPC-A-1, H1975) than in 16HBE cells cell line (Figure 1A,B). Further, we performed the IHC with a monoclonal antibody using an NSCLC tissue microarray containing 254 tumor tissues of NSCLC and 20 non-tumor tissues. Informative IHC data were obtained from 172 tumor tissues and 15 non-tumor tissues. Immunoreactivity primarily cytoplasmic presence within tumor cells and non-neoplastic bronchial or alveolar epithelial cells. There were observed staining both in tumor tissues and non-tumor tissues, but non-tumor tissues had weaker staining than tumor tissues (Independent T text, P<0.01) (Figure 1C). PACE4 positive expression in tumor cells was observed in 111 of 172 (64.53%) NSCLC (Table 2). They were further divided into 81 of 132 (61.36%) ADs, and 30 of 40 (75.00%) SCCs. There were no large cell carcinomas.
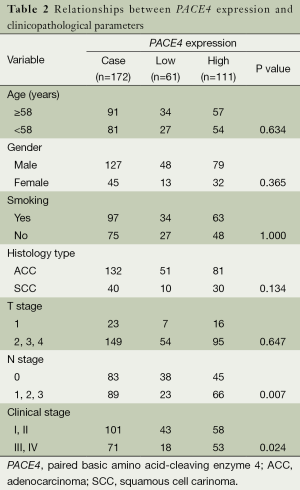
Full table
The relationship between PACE4 expression and clinicopathologic parameters
To determine whether the PACE4 expression was correlated with clinical pathological characteristics, all the samples were separated into either negative or positive PACE4 expression groups. Positive expression level of PACE4 was observed in 111/172 (64.5%) of NSCLC samples. PACE4 expression was significantly associated with lymphoid metastasis (N stage) and clinical stage (P<0.05) (Table 2) in those all NSCLC patients; However, there was no significant correlation between PACE4 expression and other clinicopathological features, such as patient age, sex, tumor sizes (T classification), smoking, and histology type (P>0.05) (Table 2).
The association of PACE4 positive expression in NSCLC with poor survival
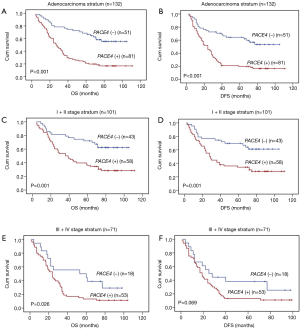
We proceeded to examine the relationship between PACE4 expression and patients’ survival after the correlation of PACE4 expression with clinicopathological features had been demonstrated. The 5-year OS and disease-free survival (DFS) rate of the informative 172 NSCLC patients were 34.7% and 34.2%, respectively (Figure 2A,B). Of these 172 NSCLC patients, Kaplan-Meier and log-rank test analyses indicated that positive PACE4 expression is significantly associated with poor overall survival (OS) (5-year survival rates, 23.1% vs. 54.5%, log-rank test, χ2=17.717, P<0.001) (Figure 2C). Similarly, PACE4 positive patients had significantly shorter DFS (5-year survival rates, 23.4% vs. 55.4%, log-rank test, χ2=20.486, P<0.001) (Figure 2D) in all the patients.
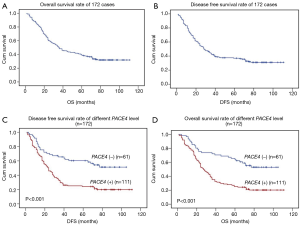
In further analyses, there was longer OS (5-year survival rates, 19.7% vs. 58.4%, log-rank test, χ2=24.806, P<0.001) (Figure 3A) and DFS (5-year survival rates, 19.5% vs. 57.1%, log-rank test, χ2=21.408, P<0.001) (Figure 3B) for the negative PACE4 expression patient in the adenocarcinoma. Then, we further to performed the clinical stratum, the result was that PACE4 positive expression was significantly associated with shorter OS (5-year survival rates, 34.4% vs. 62.5%, log-rank test, χ2=11.361, P<0.001) (Figure 3C) and DFS (5-year survival rates, 34.2% vs. 61.3%, log-rank test, χ2=10.465, P<0.001) (Figure 3D) for stage I/II patients. For stage III/IV, there was significantly associated between group positive and negative for OS (5-year survival rates, 11.3% vs. 38.2%, log-rank test, χ2=4.964, P<0.05) (Figure 3E) but not DFS (5-year survival rates, 10.8% vs. 37.6%, log-rank test, χ2=3.298, P>0.05) (Figure 3F). There was no significant difference was found between negative and positive PACE4 expression for squamous cell carcinoma patient (P>0.05, Figure not show).
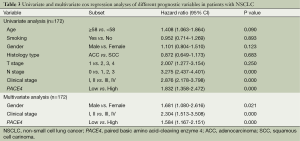
Univariate and multivariate analyses of prognostic variables in patients with NSCLC
To determine whether the PACE4 could serve as a risk factor with clinical usefulness, we examined OS using Cox regression proportional hazard analyses. The clinicopathological factors included age, gender, smoking history, histology, T stage, N stage, clinical stage. As seen in Table 3, Univariate Cox regression analyses revealed that positive expression of PACE4 was associated with a significantly increased risk of cancer-related death [hazards ratio (HR): 1.832; 95% confidence interval (CI): 1.358-2.472; P<0.001] in NSCLC patients, which indicate that PACE4 positive status increased the hazard of lung cancer-related death by two times that of PACE4 negative expression. The RRs indicated that N stage (P<0.001), clinical stage (P<0.001), were worse predictors.
Full table
After adjusting for potential confounding factors, positive expression of PACE4 in NSCLC was found to predict poorer survival in an independent manner (HR: 1.584; 95% CI: 1.167-2.151; P<0.001) (Table 3). Analyses using multivariate Cox regression model for clinicopathologic diagnoses (Table 3) gave rise to the following results: Gender (HR: 1.681; 95% CI: 1.080-2.616; P=0.021) and clinical staging (HR: 2.304; 95% CI: 1.513-3.508; P<0.001) predicted poor OS. The RRs showed no obvious differences when including other clinical parameters such as age, smoking history, histology, T stage, N stage in the analyses using the multivariate Cox regression model (P>0.05; data not shown).
Discussion
In this study, through RT-PCR and IF, there were two of the six lung cancer cells PACE4 was high expression, it is not enough to say that PACE4 was expression in NSCLC. The reason may be that most of the lung cancer cells we selected were low PACE4 expression, but by IHC and tissue microarray, we detected more than 170 cases of NSCLC tissues and it showed that PACE4 was high expression in lung cancer tissues than normal adjacent lung tissues. Studies have shown that, over-expression of PACE4 was observed in mouse basal keratinocytes, prostate and breast cancer cells (10,12,20). And high expression of PACE4 in all different clinical stages of human prostate tumor tissues (32). These results were similar to ours.
Further, we analyzed the associated with the PACE4and Clinicopathologic Parameters and survival of resected NSCLC patients. PACE4 positive expression was significantly associated with poorer OS and DFS, the decreased OS and DFS probability among clinical stage and adenocarcinoma patients, especially stage I/II subgroup and adenocarcinoma subgroup (P<0.0001). Our findings also revealed that PACE4 expression was significantly associated with clinical stage and nodal status in NSCLC. These results suggest that PACE4 expression may be important for the acquisition of migration and invasion capabilities of tumor cells, which subsequently results in poorer prognosis in patients with resected NSCLC. And how PACE4 lead to short survival time in lung cancer patients? Previous reports suggest that, over-expression in a stable manner of alpha1-PDX and the prosegment ppPACE4 in MDA-MB-231 breast cancer cells resulted in increased matrix metalloproteinase (MMP)-9 activity and a reduced secretion of tissue inhibitor of metalloproteinase 1 (TIMP-1). This was associated with significant enhancement in cell motility, migration, and invasion of collagen in vitro (33). Targeting both furin and PACE4 to the epidermis in double transgenic mice did not have an additive effect on tumor incidence/multiplicity but did enhance the tumor histopathologic grade (34). PACE4 play a direct role in sustained prostate cancer progression is most likely through the upregulated processing of growth factors or through the aberrant processing of growth factors (32). And its expression is also controlled by E-box elements in HepG2 and GH4C1 cells (35). The highest PACE4-level and related TF expression indicated of the strongest malignancy and invasiveness. It was significantly found that miR-124 was presented with the biggest odd to target PACE4-3'UTR, the capability of decreasing PACE expression and slowing down cell growth and cell invasion. It was clear that PACE4 level was closely associated with malignancy and invasiveness of PCa in vivo or in vitro. MiR-124 played a crucial role inhibiting PACE4 transcription thus exhibiting obvious effects of antiproliferation and antiaggression of PCa (36). In the present study the possible role of PACE4 as a trigger of malignant conversion by transfecting with a full-length PACE4 cDNA, three keratinocyte cell lines with no or little tumorigenic potential, invasion and MMP processing were remarkably reduced when PACE4 was inhibited with a specific antibody. PACE4 is not only able to enhance the invasive ability of malignant cells as demonstrated previously, but also played a significant role in converting non-invasive keratinocytes into malignant cells (37). Furin and PACE4 process stromelysin 3 (MMP-11 or Str-3), an MMP involved in tumor invasion, into its mature, active form. Similarly, a growing family of MMPs, known as membrane-type metalloproteinases (MT-MMPs), and growth factors and adhesion molecules such as E-cadherin show similar amino-acid motifs and thus could be activated by furin and PACE4 (38). Promoter analysis indicated that two E2F consensus binding sites (-117/-110 and -86/-79) in the 5'-flanking region of the human PACE4 gene function as positive regulatory elements. Mutation of these sites abolished PACE4 promoter response to E2F1 as well as binding of E2F1 in electrophoretic mobility-shift assays. Other E2F members, E2F2 and E2F3, also activated PACE4 expression, as in the case of E2F1. These results indicate a novel mechanism for E2F family-mediated promotion of carcinoma invasiveness through PACE4 (39). PACE4 overexpression and activity accounts for an increased proliferation rate, exaggerated sensitivity to the PC inhibitor CMK, and interference with IGF-1R autophosphorylation. Mice overexpressing PACE4 exhibited tumors of increased growth rate and invasive potential when exposed to the human carcinogen B(a)P, further supporting the significance of PCs in tumor growth and progression (40). We hypothesized whether it is through these mechanisms that PACE4 to regulate the lung cancer, it is our future research.
In conclusion, we have provided the first clinical evidence that PACE4 is expressed in a subset of NSCLC and its expression is related to clinicopathological factors. We demonstrated that PACE4 expression is a prognostic indicator of poor survival among patients with resected NSCLC, although its prognostic significance still requires confirmation with larger patient populations.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 8120184).
Authors’ contributions: YJ Zhang designed the overall study and edited the paper. QN Wu and YE Lin designed and carried out the IHC of the experiments, collected and analyzed data, and cowrote the paper. XD Lin and GQ Li carried out RT-PCR and IF of the experiments, XD Lin collected with QN Wu and YE Lin analyzed the data.
Disclosure: The authors declare no conflict of interest.
References
- Osarogiagbon RU, Allen JW, Farooq A, et al. Pathologic lymph node staging practice and stage-predicted survival after resection of lung cancer. Ann Thorac Surg 2011;91:1486-92. [PubMed]
- Smeekens SP. Processing of protein precursors by a novel family of subtilisin-related mammalian endoproteases. Biotechnology (N Y) 1993;11:182-6. [PubMed]
- Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov 2012;11:367-83. [PubMed]
- Couture F, D'Anjou F, Day R. On the cutting edge of proprotein convertase pharmacology: from molecular concepts to clinical applications. Biomol Concepts 2011;2:421-38. [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [PubMed]
- Fu J, Bassi DE, Zhang J, et al. Transgenic overexpression of the proprotein convertase furin enhances skin tumor growth. Neoplasia 2012;14:271-82. [PubMed]
- Dogar AM, Towbin H, Hall J. Suppression of latent transforming growth factor (TGF)-beta1 restores growth inhibitory TGF-beta signaling through microRNAs. J Biol Chem 2011;286:16447-58. [PubMed]
- Mahloogi H, Bassi DE, Klein-Szanto AJ. Malignant conversion of non-tumorigenic murine skin keratinocytes overexpressing PACE4. Carcinogenesis 2002;23:565-72. [PubMed]
- McColl BK, Paavonen K, Karnezis T, et al. Proprotein convertases promote processing of VEGF-D, a critical step for binding the angiogenic receptor VEGFR-2. FASEB J 2007;21:1088-98. [PubMed]
- Bassi DE, Lopez De Cicco R, Cenna J, et al. PACE4 expression in mouse basal keratinocytes results in basement membrane disruption and acceleration of tumor progression. Cancer Res 2005;65:7310-9. [PubMed]
- Page RE, Klein-Szanto AJ, Litwin S, et al. Increased expression of the pro-protein convertase furin predicts decreased survival in ovarian cancer. Cell Oncol 2007;29:289-99. [PubMed]
- Cheng M, Watson PH, Paterson JA, et al. Pro-protein convertase gene expression in human breast cancer. Int J Cancer 1997;71:966-71. [PubMed]
- Khatib AM, Siegfried G, Prat A, et al. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J Biol Chem 2001;276:30686-93. [PubMed]
- Mercapide J, Lopez De Cicco R, Bassi DE, et al. Inhibition of furin-mediated processing results in suppression of astrocytoma cell growth and invasiveness. Clin Cancer Res 2002;8:1740-6. [PubMed]
- Cheng M, Watson PH, Paterson JA, et al. Pro-protein convertase gene expression in human breast cancer. Int J Cancer 1997;71:966-71. [PubMed]
- Singh H, Heng S, Nicholls PK, et al. Proprotein convertases in post-menopausal endometrial cancer: distinctive regulation and non-invasive diagnosis. Biochem Biophys Res Commun 2012;419:809-14. [PubMed]
- Kiefer MC, Tucker JE, Joh R, et al. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol 1991;10:757-69. [PubMed]
- Tsuji A, Hine C, Tamai Y, et al. Genomic organization and alternative splicing of human PACE4 (SPC4), kexin-like processing endoprotease. J Biochem 1997;122:438-52. [PubMed]
- Wang F, Wang L, Pan J. PACE4 regulates proliferation, migration and invasion in human breast cancer MDA-MB-231 cells. Mol Med Rep 2015;11:698-704. [PubMed]
- Couture F, D'Anjou F, Desjardins R, et al. Role of proprotein convertases in prostate cancer progression. Neoplasia 2012;14:1032-42. [PubMed]
- D’Anjou F, Couture F, Desjardins R, et al. Knockdown strategies for the study of proprotein convertases and proliferation in prostate cancer cells. Methods Mol Biol 2014;1103:67-82. [PubMed]
- Hubbard FC, Goodrow TL, Liu SC, et al. Expression of PACE4 in chemically induced carcinomas is associated with spindle cell tumor conversion and increased invasive ability. Cancer Res 1997;57:5226-31. [PubMed]
- Kim H, Tabata A, Tomoyasu T, et al. Estrogen stimuli promote osteoblastic differentiation via the subtilisin-like proprotein convertase PACE4 in MC3T3-E1 cells. J Bone Miner Metab 2015;33:30-9. [PubMed]
- Longuespée R, Couture F, Levesque C, et al. Implications of Proprotein Convertases in Ovarian Cancer Cell Proliferation and Tumor Progression: Insights for PACE4 as a Therapeutic Target. Transl Oncol 201. [Epub ahead of print]. [PubMed]
- Bando M, Matsuoka A, Tsuji A, et al. The proprotein convertase PACE4 is upregulated by PDGF-BB in megakaryocytes: gene expression of PACE4 and furin is regulated differently in Dami cells. J Biochem 2002;132:127-34. [PubMed]
- Yuasa K, Masuda T, Yoshikawa C, et al. Subtilisin-like proprotein convertase PACE4 is required for skeletal muscle differentiation. J Biochem 2009;146:407-15. [PubMed]
- Yuasa K, Futamatsu G, Kawano T, et al. Subtilisin-like proprotein convertase paired basic amino acid-cleaving enzyme 4 is required for chondrogenic differentiation in ATDC5 cells. FEBS J 2012;279:3997-4009. [PubMed]
- Bassi DE, Zhang J, Cenna J, et al. Proprotein convertase inhibition results in decreased skin cell proliferation, tumorigenesis, and metastasis. Neoplasia 2010;12:516-26. [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [PubMed]
- Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. Natl Compr Canc Netw 2013;11:645-53; quiz 653.
- Finn RS, Press MF, Dering J, et al. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol 2009;27:3908-15. [PubMed]
- D’Anjou F, Routhier S, Perreault JP, et al. Molecular Validation of PACE4 as a Target in Prostate Cancer. Transl Oncol 2011;4:157-72. [PubMed]
- Lapierre M, Siegfried G, Scamuffa N, et al. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells’ malignant phenotypes: role of tissue inhibitors of metalloproteinase-1. Cancer Res 2007;67:9030-4. [PubMed]
- Fu J, Bassi DE, Zhang J, et al. Enhanced UV-induced skin carcinogenesis in transgenic mice overexpressing proprotein convertases. Neoplasia 2013;15:169-79. [PubMed]
- Tsuji A, Yoshida S, Hasegawa S, et al. Human subtilisin-like proprotein convertase, PACE4 (SPC4) gene expression is highly regulated through E-box elements in HepG2 and GH4C1 cells. J Biochem 1999;126:494-502. [PubMed]
- Kang S, Zhao Y, Hu K, et al. miR-124 exhibits antiproliferative and antiaggressive effects on prostate cancer cells through PACE4 pathway. Prostate 2014;74:1095-106. [PubMed]
- Mahloogi H, Bassi DE, Klein-Szanto AJ. Malignant conversion of non-tumorigenic murine skin keratinocytes overexpressing PACE4. Carcinogenesis 2002;23:565-72. [PubMed]
- Bassi DE, Mahloogi H, Klein-Szanto AJ. The proprotein convertases furin and PACE4 play a significant role in tumor progression. Mol Carcinog 2000;28:63-9. [PubMed]
- Yuasa K, Suzue K, Nagahama M, et al. Transcriptional regulation of subtilisin-like proprotein convertase PACE4 by E2F: possible role of E2F-mediated upregulation of PACE4 in tumor progression. Gene 2007;402:103-10. [PubMed]
- Bassi DE, Cenna J, Zhang J, et al. Enhanced aggressiveness of benzopyrene-induced squamous carcinomas in transgenic mice overexpressing the proprotein convertase PACE4 (PCSK6). Mol Carcinog 2014. [Epub ahead of print]. [PubMed]


