EPHX1 rs2234922 polymorphism and lung cancer susceptibility in Asian populations: a meta-analysis
Introduction
Lung cancer remains the deadliest cancer worldwide despite improvements in diagnostic and therapeutic techniques (1). Its incidence has yet to peak in many parts of world, particularly in China, which has become a major public health challenge (2). The mechanism of lung carcinogenesis is still not fully understood. Besides smoking status established as the most important single factor in causing lung cancer, host factors, including genetic polymorphisms, have been some growing interest in the study of the tumorigenesis of lung cancer (3). Many environmental carcinogens require metabolic activation by various drug-metabolizing enzymes.
Microsomal epoxide hydrolase 1 (EPHX1) plays an important role in both the activation and detoxification of PAHs and aromatic amines (4,5). The human EPHX1 gene is 35.48 kb with nine exons and eight introns on chromosome 1q42.1. There are more than 100 single-nucleotide polymorphisms in EPHX1 gene. A dominant polymorphism of A > G (His139Arg) in exon 4 of EPHX1 has been shown to influence enzyme activity. It has been demonstrated that a variant Arg allele at codon 139 (EPHX1 His139Arg) increases enzyme activity by at least 25% (high EPHX1 activity allele) (6). The predominant homozygous allele, the heterozygous allele and the homozygous rare allele of the EPHX1 rs2234922 polymorphism are named the homozygous wild-type genotype (A/A), G allele carriers (A/G) and the rare homozygote (G/G), respectively.
Many studies have investigated associations between EPHX1 rs2234922 gene polymorphism and lung cancer risk, but there is the perception that the findings have been inconsistent. A single study may be too underpowered to detect a possible small effect of the polymorphisms on lung cancer, especially when the sample size is relatively small. Different types of study populations may also contribute the disparate findings. Hence, we performed a meta-analysis of all eligible studies to derive a more precise estimation of the associations of EPHX1 rs2234922 polymorphism with lung cancer.
Materials and methods
Publication search
The electronic databases PubMed were searched for studies to include in the present meta-analysis, using the terms: “EPHX1” or “microsomal epoxide hydrolase 1”, “rs2234922”, “polymorphism” and “lung cancer”. An upper date limit of Oct 01, 2014 was applied; no lower date limit was used. The search was performed without any restrictions on language and was focused on studies that had been conducted in humans. Concurrently, the reference lists of reviews and retrieved articles were searched manually. Only full-text articles were included. When the same patient population appeared in several publications, only the most recent or complete study was included in this meta-analysis.
Inclusion criteria
The included studies have to meet the following criteria: (I) evaluating the EPHX1 rs2234922 polymorphism and lung cancer risk; (II) case-control studies; and (III) supply the number of individual genotypes for EPHX1 rs2234922 genotype in lung cancer cases and controls, respectively.
Data extraction
Information was carefully extracted from all eligible publications independently by two authors according to the inclusion criteria listed above. Disagreement was resolved by discussion between the two authors.
The following data were collected from each study: first author’s surname, year of publication, ethnicity, total numbers of cases and controls, and numbers of cases and controls with the AA, AG and GG genotypes, respectively. If data from any of the above categories were not reported in the primary study, items were treated as “not applicable”. We did not contact the author of the primary study to request the information. Different ethnicity descents were categorized as Asian, and non-Asian. We did not require a minimum number of patients for a study to be included in our meta-analysis.
Statistical analysis
Odds ratio (OR) with 95% confidence interval (CI) was used to assess the strength of association between the EPHX1 rs2234922 polymorphism and lung cancer risk. The pooled ORs for the risk associated with the genotypes of homozygote G/G and G allele carriers (A/G + G/G) with the A/A genotype were calculated. Heterogeneity assumption was checked by the chi-square-based Q-test (7). A P value greater than 0.10 for the Q-test indicates a lack of heterogeneity among studies, so the pooled OR estimate of the each study was calculated by the fixed-effects model (the Mantel-Haenszel method) (8). Otherwise, the random-effects model (the DerSimonian and Laird method) was used (9). One-way sensitivity analyses were performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled OR (10). An estimate of potential publication bias was carried out by the funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure the funnel plot asymmetry on the natural logarithm scale of the OR. The significance of the intercept was determined by the t-test suggested by Egger (P<0.05 was considered representative of statistically significant publication bias) (11). All of the calculations were performed using STATA version 11.0 (Stata Corporation, College Station, TX).
Results
Study characteristics
A total of eight publications involving 1,175 lung cancer cases and 1,550 controls met the inclusion criteria and were ultimately analyzed (12-19). Table 1 presents the main characteristics of these studies. Among the eight publications, all were published in English. The sample sizes ranged from 69 to 216. Almost all of the cases were histologically confirmed. Controls were mainly healthy populations. There were two groups of Indian, one group of Japan and five groups of China.

Full table
Meta-analysis results
Overall, for the homozygote G/G and G allele carriers (A/G + G/G), the pooled ORs for all studies combined 1,175 lung cancer cases and 1,550 controls were 1.47 (95% CI: 1.18-1.79, P=0.007 for heterogeneity) and 1.36 (95% CI: 1.14-1.62, P=0.828 for heterogeneity) (Figure 1), when compared with the homozygous wild-type genotype (A/A).
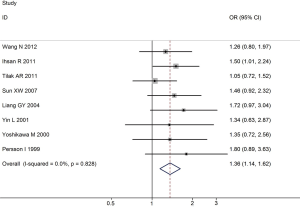
Sensitivity analyses
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs, and the corresponding pooled ORs were not materially altered (data not shown).
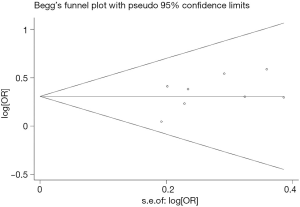
Publication bias
Begg’s funnel plot and Egger’s test were performed to access the publication bias of literatures. Evaluation of publication bias for A/G + G/G versus AA showed that the Egger test was not significant (P=0.202). The funnel plots for publication bias (Figure 2) also did not show some asymmetry. These results did not indicate a potential for publication bias.
Discussion
The expression of EPHX1 has been detected in microsome, endoplasmic reticulum, and integral membrane, especially in the lung, liver, and kidney tissues (20). EPHX1 plays an important role in both the activation and detoxification of many carcinogens, such as polycyclic aromatic hydrocarbons and aromatic amines (6,21). The polymorphism at exon 4 of the EPHX1 gene has been reported to be associated with variations in EPHX1 activity (21-23). Individuals carrying different genotypes of EPHX1 are different in susceptibility to a number of carcinogen-induced cancers, which is attributed to variations in the enzymatic activity of EPHX1 as a result of such polymorphisms (24,25).
The present meta-analysis investigating the relationship between EPHX1 rs2234922 polymorphism and lung cancer risk provided the most comprehensive evidence. Significant association of EPHX1 rs2234922 polymorphic and lung cancer risk was demonstrated. Meta-analyses of total studies showed that EPHX1 rs2234922 polymorphism was associated with lung cancer risk for the homozygote G/G and G allele carriers (A/G + G/G) when compared with the homozygous wild-type genotype (A/A). Sensitivity analyses by sequential omission of any individual studies and subgroup analyses by case sample size and ethnicity suggested the findings were highly unlikely due to chance. Publication bias analysis demonstrated that bias favoring publications of positive studies was inexistent.
Some limitations of this meta-analysis should be acknowledged. Firstly, heterogeneity is a potential problem when interpreting all the results of meta-analyses. Although we minimized the likelihood by performing a careful search for published studies, using the explicit criteria for study inclusion, performing data extraction and data analysis strictly, the significant between-study heterogeneity still existed in almost each comparison. The presence of heterogeneity can result from differences in the selection of controls, age distribution, lifestyle factors and so on. Although most of the controls were selected from healthy populations, some studies had selected controls among friends or family of lung cancer patients or patients with other diseases. Secondly, only published studies were included in this meta-analysis. The presence of publication bias indicates that non-significant or negative findings may be unpublished. Lastly, our results were based on unadjusted estimates, while a more precise analysis should be conducted if individual data were available, which would allow for the adjustment by other covariates including age, ethnicity, family history, environmental factors and lifestyle.
In conclusion, this meta-analysis suggests that the EPHX1 rs2234922 polymorphism is associated with lung cancer risk among Asian population. In addition, it is necessary to conduct large trials using standardized unbiased methods, homogeneous lung cancer patients and well matched controls, with the assessors blinded to the data.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Gulati S, Mulshine JL. Lung cancer screening guidelines: common ground and differences. Transl Lung Cancer Res 2014;3:131-8. [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [PubMed]
- Toh CK, Gao F, Lim WT, et al. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol 2006;24:2245-51. [PubMed]
- Decker M, Arand M, Cronin A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch Toxicol 2009;83:297-318. [PubMed]
- Arand M, Cronin A, Oesch F, et al. The telltale structures of epoxide hydrolases. Drug Metab Rev 2003;35:365-83. [PubMed]
- Benhamou S, Reinikainen M, Bouchardy C, et al. Association between lung cancer and microsomal epoxide hydrolase genotypes. Cancer Res 1998;58:5291-3. [PubMed]
- Cochran WG. The combination of estimates from different experiments. Biometrics 1954;10:101-29.
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719-48. [PubMed]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88. [PubMed]
- Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [PubMed]
- Bhat IA, Pandith AA, Bhat BA, et al. Lack of association of a common polymorphism in the 3' -UTR of interleukin 8 with non small cell lung cancer in Kashmir. Asian Pac J Cancer Prev 2013;14:4403-8. [PubMed]
- Wang N, Wu Y, Zhou X, et al. Association between genetic polymorphism of metabolizing enzymes and DNA repairing enzymes and the susceptibility of lung cancer in Henan population. Wei Sheng Yan Jiu 2012;41:251-6. [PubMed]
- Ihsan R, Chauhan PS, Mishra AK, et al. Multiple analytical approaches reveal distinct gene-environment interactions in smokers and non smokers in lung cancer. PLoS One 2011;6:e29431. [PubMed]
- Tilak AR, Kumar S, Jain M, et al. Association of functionally important polymorphism of microsomal epoxide hydrolase gene (EPHX1) with lung cancer susceptibility. Cancer Invest 2011;29:411-8. [PubMed]
- Sun XW, Ma YY, Wang B. The interaction between microsomal epoxide hydrolase polymorphisms and indoor pollution in non small cell lung cancer. Zhonghua Yu Fang Yi Xue Za Zhi 2007;41 Suppl:30-4. [PubMed]
- Liang GY, Pu YP, Yin LH. Studies of the genes related to lung cancer susceptibility in Nanjing Han population, China. Yi Chuan 2004;26:584-8. [PubMed]
- Yin L, Pu Y, Liu TY, et al. Genetic polymorphisms of NAD(P)H quinone oxidoreductase, CYP1A1 and microsomal epoxide hydrolase and lung cancer risk in Nanjing, China. Lung Cancer 2001;33:133-41. [PubMed]
- Yoshikawa M, Hiyama K, Ishioka S, et al. Microsomal epoxide hydrolase genotypes and chronic obstructive pulmonary disease in Japanese. Int J Mol Med 2000;5:49-53. [PubMed]
- Persson I, Johansson I, Lou YC, et al. Genetic polymorphism of xenobiotic metabolizing enzymes among Chinese lung cancer patients. Int J Cancer 1999;81:325-9. [PubMed]
- Omiecinski CJ, Aicher L, Holubkov R, et al. Human peripheral lymphocytes as indicators of microsomal epoxide hydrolase activity in liver and lung. Pharmacogenetics 1993;3:150-8. [PubMed]
- Jang JH, Cotterchio M, Borgida A, et al. Genetic variants in carcinogen-metabolizing enzymes, cigarette smoking and pancreatic cancer risk. Carcinogenesis 2012;33:818-27. [PubMed]
- Brighenti M. MicroRNA and MET in lung cancer. Ann Transl Med 2015;3:68. [PubMed]
- Okada M, Mimae T, Tsutani Y, et al. Segmentectomy versus lobectomy for clinical stage IA lung adenocarcinoma. Ann Cardiothorac Surg 2014;3:153-9. [PubMed]
- Kiyohara C, Yoshimasu K, Takayama K, et al. EPHX1 polymorphisms and the risk of lung cancer: a HuGE review. Epidemiology 2006;17:89-99. [PubMed]
- Sivonova MK, Dobrota D, Matakova T, et al. Microsomal epoxide hydrolase polymorphisms, cigarette smoking and prostate cancer risk in the Slovak population. Neoplasma 2012;59:79-84. [PubMed]


