Reduced expression of circRNA novel_circ_0005280 and its clinical value in the diagnosis of non-small cell lung cancer
Introduction
Lung cancer is currently the leading cause of cancer-associated mortality worldwide (1-3). Primary lung cancer is classified as small cell lung carcinoma (SCLC) and non-SCLC (NSCLC). NSCLC, which originates from non-small cells in the lung, accounts for 83% of all lung cancer cases and has high mortality (4). The 5-year survival rate of patients with NSCLC is very low, as patients are often in the mid or advanced stage when they are diagnosed (2,4). Therefore, approaches for preventing and treating NSCLC are needed. It is also important to develop methods for the early diagnosis and accurate prognosis of NSCLC.
Circular RNAs (circRNAs) are endogenous non-coding RNAs that have become a research hotspot. CircRNAs were originally described in the late 1970s and are produced by back-splicing of a single pre-mRNA. These molecules have no 5' or 3' ends, and tend to form ring structures (4-6). The conservation and stability of circRNAs and their degradation mode make them important candidates for biomarker discovery in disease research (7-11). Many studies have discovered that circRNAs are widespread, abundant, expressed across species, and are involved in physiological and pathological processes (12-15). Increasingly, studies have found that they play an important role in the occurrence and development of tumors. For example, some circRNAs are associated with hepatocellular carcinoma, glioma, breast cancer, gastric cancer, and leukemia (16-23). Competing endogenous RNA, the relationship between circRNAs and exosomes, the interaction between circRNAs and proteins, and the degradation mode of circRNAs have been the focus of many researchers.
Notably, through the continuous development of bioinformatics analysis and high-throughput sequencing technology, many circRNAs have been reported as potential biomarkers for cancer diagnosis or prognosis (10,11,24,25). Our RNAsequencing (RNA-seq) results revealed a novel_circ_0005280, which is 525 nucleotides in length, is transcribed from chr2 166447611–166472445, and is composed of 4 exons (166447612–166447708, 166456870–166457076, 166470615–166470706, and 166472317–166472445); its associated gene symbol is SCN7A. SCN7A (Sodium Voltage-Gated Channel Alpha Subunit 7) is a Protein Coding gene. Diseases associated with SCN7A include Periodic Paralysis and Erythromelalgia. Among its related pathways are Neuroscience and Activation of cAMP-Dependent PKA. Gene Ontology (GO) annotations related to this gene include ion channel activity and sodium channel activity (GeneCards summary for SCN7A Gene).
To the best of our knowledge, the present study is the first to evaluate the role of novel_circ_0005280 in NSCLC. Our results show that the novel_circ_0005280 has high sensitivity and specificity in the diagnosis of non-small cell lung cancer, which has not been seen in other similar studies. Therefore, we think that novel_circ_0005280 plays an important role in the occurrence of NSCLC and may be useful for the diagnosis and prognosis of NSCLC.
We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2977).
Methods
Patients and clinical information
Clinical NSCLC tissue samples were collected from patients at Shengjing Hospital of China Medical University from July 2017 to January 2018. The inclusion criteria were as follows: (I) patients diagnosed with primary NSCLC and without other malignancies; (II) availability of complete medical and follow-up records; and (III) fresh tumor tissues and adjacent normal tissues preserved in liquid nitrogen immediately after surgery. Lung cancer tissue was available from the tumor center; adjacent tissue was collected from an area >5 cm beyond the edge of the carcinoma. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All procedures were approved by the Ethics Committee of Shengjing Hospital of China Medical University (certificate No. 2017PS339K), and all participants provided signed informed consent.
Total RNA-seq and screening of circRNAs
In the screening stage, 3 pairs of cancerous and adjacent normal tissues were collected from patients with NSCLC during appointments at Shengjing Hospital of China Medical University; these were used for total RNA-seq. Sequencing was performed by Beijing Nova Biotechnology (Beijing, China) using a HiSeq 2000 (Illumina, San Diego, CA, USA) instrument. A quality assessment of the original sequencing data was carried out, and the sequencing data were compared to the reference gene using TopHat software (version 2.1.1). Expression levels were evaluated as fragments per kilobase per million mapped reads (FPKM). The Cufflinks method was used to quantify gene expression values and screen for differential genes. The screening criterion of differential genes in the present study was a false discovery rate <0.05. The gene expression value was evaluated as log10 (FPKM +1).
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
According to the RNA-seq results, bioinformatics analysis was performed to select candidate circRNAs. Tumor tissues and 27 adjacent tissues from 41 patients with NSCLC were collected and further verified with real-time qRT-PCR. TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to separate total RNA from tissues for the quantitative detection of circRNA, according to the manufacturer’s instructions. The Geneseed II First Strand cDNA Synthesis Kit (Geneseed, Guangzhou, China) was used to synthesize cDNA. First-strand cDNA synthesis reaction was carried out under the following conditions: 25 °C for 10 min, 42 °C for 15 min, and 85 °C for 5 min. qRT-PCR was performed using an ABI 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using Geneseed qPCR SYBR Green Master Mix (Geneseed, Guangzhou, China). The qPCR reaction program was set as follows: stage 1, pre-denaturation, reps: 1, 95 °C for 5 min; stage 2, cyclic reaction, reps: 40, 95 °C for 10 s, 60 °C for 34 s; and stage 3, melting curve, reps: 1, 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. The primer sequences used for the PCR are shown in Table 1. All experiments were repeated 3 times. Relative gene expression levels were calculated using the 2-ΔΔCt method, where ΔΔCt = ΔCt (carcinoma sample) − ΔCt (adjacent normal sample).

Full table
Statistical analysis
The Mann-Whitney and Wilcoxon matched-pair tests were used to compare the expression level of novel_circ_0005280 between NSCLC tissues and adjacent non-tumorous tissues. Pearson’s χ2-test was used to evaluate the relationship between the expression of novel_circ_0005280 and the clinicopathological parameters of NSCLC. A receiver-operating characteristic (ROC) curve was used to evaluate the diagnostic value of this circRNA. The log-rank test was performed with Kaplan-Meier survival analysis. Analyses were performed using SPSS (version 24.0; SPSS, Chicago, IL, USA) and GraphPad Prism software (version 7.0; GraphPad, La Jolla, CA, USA). Statistical significance was set at P<0.05.
Results
RNA-seq
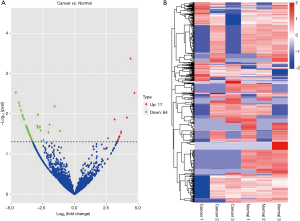
High-throughput sequencing can estimate the expression profiles of circRNA genes. Two groups of patients were screened for the differential expression of genes. A total of 2,573 circRNAs were detected by RNA-seq in 3 pairs of samples. CircRNA expression was considered significantly upregulated or downregulated at fold-changes of >2.0 or <–2.0, respectively, with P≤0.05. Among these circRNAs, 17 and 64 circRNAs showed higher and lower expressions, respectively. The distribution of the differentially expressed genes is represented by a volcano plot (Figure 1A).
Differentially expressed circRNAs were analyzed by hierarchical clustering based on the log10 (transcripts per million, TPM +1) values of circRNAs, and circRNAs with the same or similar expression patterns were clustered and displayed in a heat map (Figure 1B).
qRTPCR and Sanger sequencing
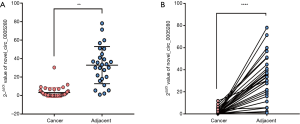
Four circRNAs (has_circ_0067582, novel_circ_0005280, has_circ_0005692, and novel_circ_0008866) were randomly selected for qRT-PCR validation and Sanger sequencing to determine the reliability of the RNA-seq results. The results revealed significantly different expression of novel_circ_0005280 in cancer tissues and adjacent tissues (Figure 2). The RNAs were directly sequenced from RT-PCR fragments to obtain the consensus sequence using a traditional Sanger sequencing approach and the circBase database for circular RNAs (www.circbase.org) (Figure 3).
Relationship between hsa_circ_0005280 levels and clinicopathological factors
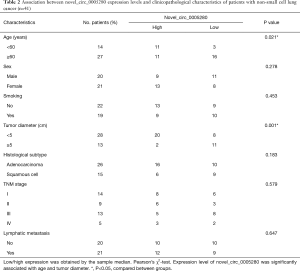
We investigated whether the expression levels of novel_circ_0005280 were associated with the available clinicopathological features of patients with NSCLC (Table 2), and found a correlation between novel_circ_0005280 expression and age (P=0.021) and tumor size (P=0.001) in patients with NSCLC; however, there was no correlation with smoking, sex, or other factors.
Full table
ROC curve and the Kaplan-Meier curve of novel_circ_0005280
To evaluate the diagnostic value of novel_circ_0005280, ROC curves were plotted, and the area under the curve (AUC) was calculated (Figure 4A). The AUC of novel_circ_0005280 reached up to 0.944 (95% confidence interval: 0.890–0.998, P<0.0001). When the cutoff value was set to 10.23, the sensitivity and specificity of novel_circ_0005280 were 85.2% and 95.1%, respectively.
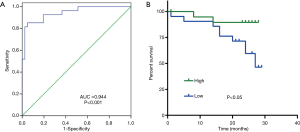
The Kaplan-Meier curve showed that patients with low novel_circ_0005280 expression had a shorter survival time than those with high expression (Figure 4B).
Discussion
Lung cancer development involves many molecular abnormalities, which are accompanied by complex biologic processes. The expression of non-coding RNA is considered a useful metric for describing the molecular characteristics of lung cancer. Many studies have suggested that microRNAs or long non-coding RNAs are related to lung cancer (26-28). These studies give us some inspiration to study circRNAs: circRNA can combine with miRNA, so maybe circRNA can have some effects on lung cancer through miRNA. And circRNA can be regarded as a special class of lncRNA, so circRNA can also have some effects on lung cancer, just like lncRNA. Various new functions of circRNAs have been discovered, such as their involvement in translation through the circRNA-protein interaction (13,29). Many circRNAs have been found to be related to the development, invasion, metastasis, or prognosis of various cancers. Although some circRNAs have been studied in lung cancer, the mechanism of the tumorigenesis of lung cancer and the practical role of circRNAs in the clinical diagnosis and treatment of NSCLC remain unclear (17,18,30-32). Relevant research is still in its infancy, and further studies are necessary.
The findings of the present study showed that novel_circ_0005280 was significantly downregulated in NSCLC tissues, demonstrating its potential as a new biomarker for the diagnosis and prognosis of NSCLC. We first performed RNA-seq analysis to determine the expression profile of circRNA in lung cancer tissues and adjacent tissues of 3 patients with human NSCLC. Our results showed that there was a significant difference in the circRNA expression between cancer and non-cancer tissues of NSCLC patients (n=3) (Figure 1). After validating the expression of 4 circRNAs in additional tissue samples, novel_circ_0005280 was found to be significantly downregulated in NSCLC (Figure 2). This is consistent with RNA-seq trends. Additionally, our experimental results showed that some clinical and pathological factors of NSCLC patients are significantly related to the low expression of novel_circ_0005280 in NSCLC (Table 2), suggesting that this circRNA can be used in the prognosis of NSCLC. The expression of novel_circ_0005280 is significantly related to the age of patients and tumor size, but we know that the older the tumor patients are, the worse the prognosis of patients with larger tumor diameter is. Therefore, we consider that the expression of novel_circ_0005280 is closely related to the prognosis of patients. Moreover, ROC curve analysis revealed the excellent discriminative capability of novel_circ_0005280, as indicated by the high AUC value (>0.90). In summary, our results suggest that novel_circ_0005280 is involved in the occurrence, development, and metastasis of NSCLC, and may be useful as a biomarker for the diagnosis and prognosis of NSCLC, which could lead to improvements in the treatment of NSCLC.
The association between circRNAs and some clinicopathological features has strong clinical value. Therefore, early cancer diagnosis can improve the 5-year survival rate of patients (10,25). To the best of our knowledge, the present stud was the first to identify the role of novel_circ_0005280 in NSCLC. Our research results showed that the expression levels of novel_circ_0005280 were lower in NSCLC tissues compared to those in paracancerous non-cancerous tissues. Novel_circ_0005280 levels in NSCLC tissues were significantly related to tumor diameter and age. Our results showed that the expression levels of novel_circ_0005280 in human NSCLC tissues were significantly correlated with the tumor diameter and age of the patient. Tumor size was positively related with tumor stage; additionally, a lower expression of novel_circ_0005280 was associated with a larger tumor diameter. Therefore, novel_circ_0005280 could be an excellent prognostic marker. Furthermore, older patients showed a lower expression of novel_circ_0005280 (Table 2), supporting this finding. Novel_circ_0005280 may be valuable for auxiliary diagnosis, as the AUC expression level of novel_circ_0005280 was 0.944, and the sensitivity and specificity were higher than those of most diagnostic markers used for lung cancer.
Conclusions
Novel_circ_0005280 may be used as a biomarker for NSCLC diagnosis and prognosis. However, the sample size in the present study was small. Further studies with larger sample sizes and in vitro experiments are needed to verify our results. Our findings provide a better understanding of the role of circRNAs in human NSCLC.
Acknowledgments
We thank Novartis Biotechnology (Beijing, China) for providing the transcriptome sequencing and subsequent bioinformatics analysis.
Funding: The present study was supported by the Shenyang Science and Technology Plan Project (No. 19-112-4-106).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2977
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2977
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2977). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All procedures were approved by the Ethics Committee of Shengjing Hospital of China Medical University (certificate No. 2017PS339K), and all participants provided signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Qian F, Yang W, Chen Q, et al. Screening for early stage lung cancer and its correlation with lung nodule detection. J Thorac Dis 2018;10:S846-59. [Crossref] [PubMed]
- Shankar A, Saini D, Dubey A, et al. Feasibility of lung cancer screening in developing countries: challenges, opportunities and way forward. Transl Lung Cancer Res 2019;8:S106-21. [Crossref] [PubMed]
- Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin 2011;61:91-112. [Crossref] [PubMed]
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep 2015;10:103-11. [Crossref] [PubMed]
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Guo Y, Wei X, Peng Y. Structure-Mediated Degradation of CircRNAs. Trends Cell Biol 2020;30:501-3. [Crossref] [PubMed]
- Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: Biomarkers and master regulators. Semin Cancer Biol 2019;58:90-9. [Crossref] [PubMed]
- Di X, Jin X, Li RW, et al. CircRNAs and lung cancer: Biomarkers and master regulators. Life sciences 2019;220:177-85. [Crossref] [PubMed]
- Li X, Ding J, Wang X, et al. NUDT21 regulates circRNA cyclization and ceRNA crosstalk in hepatocellular carcinoma. Oncogene 2020;39:891-904. [Crossref] [PubMed]
- Du WW, Zhang C, Yang W, et al. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017;7:4183-91. [Crossref] [PubMed]
- Feng J, Chen K, Dong X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer 2019;18:35. [Crossref] [PubMed]
- Li J, Sun D, Pu W, et al. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020;6:319-36. [Crossref] [PubMed]
- Hu ZQ, Zhou SL, Li J, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020;72:906-22. [Crossref] [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell 2019;176:869-881.e13. [Crossref] [PubMed]
- Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer 2017;16:25. [Crossref] [PubMed]
- Ding C, Yi X, Wu X, et al. Exosome-mediated transfer of circRNACircNFIX enhances temozolomide resistance in glioma. Cancer Lett 2020;479:1-12. [Crossref] [PubMed]
- Smid M, Wilting SM, Uhr K, et al. The circular RNome of primary breast cancer. Genome Res 2019;29:356-66. [Crossref] [PubMed]
- Li R, Jiang JJ, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci 2020;77:1661-80. [Crossref] [PubMed]
- Wang S, Zhang X, Li Z, et al. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. Oncogene 2019;38:6985-7001. [Crossref] [PubMed]
- Wu Z, Sun H, Wang C, et al. Mitochondrial Genome-Derived circRNA mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol Ther Nucleic Acids 2020;20:801-11. [Crossref] [PubMed]
- Chen L, Wang C, Sun H, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 2020. Epub ahead of print. [Crossref] [PubMed]
- Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. International journal of medical sciences 2019;16:292-301. [Crossref] [PubMed]
- Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM, et al. Somatic Mutations in miRNA Genes in Lung Cancer-Potential Functional Consequences of Non-Coding Sequence Variants. Cancers (Basel) 2019;11:793. [Crossref] [PubMed]
- Seiler J, Breinig M, Caudron-Herger M, et al. The lncRNA VELUCT strongly regulates viability of lung cancer cells despite its extremely low abundance. Nucleic Acids Res 2017;45:5458-69. [Crossref] [PubMed]
- Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett 2016;371:99-106. [Crossref] [PubMed]
- Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol 2020;22:2162-9. [Crossref] [PubMed]
- Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine 2019;44:182-93. [Crossref] [PubMed]
- Zhou J, Zhang S, Chen Z, et al. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis 2019;10:885. [Crossref] [PubMed]
- Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med 2018;7:2783-91. [Crossref] [PubMed]
(English Language Editor: R. Scott)