Consistency of genotyping data from simultaneously collected plasma circulating tumor DNA and tumor-DNA in lung cancer patients
Introduction
In recent years, tumor genotype data has become indispensable during treatment of advanced lung cancer. It is commonly used in strategic decision-making when considering whether targeted therapy, immunotherapy, or traditional chemotherapy should be administered (1-3). Hence, the collection of accurate and extensive molecular tumor information has important clinical implications.
Since the discovery by Mandel 1964 of free nucleic acid molecules in the plasma (4), circulating tumor DNA (ctDNA) has led to several research breakthroughs. Early detection, treatment selection, efficacy prediction, and recurrence monitoring of cancer have been studied using ctDNA, which has enabled the development of the liquid biopsy approach. Liquid biopsy is considered a promising clinical tool due to its advantages, which include being noninvasive and highly reproducible. It is also able to overcome the shortcomings of tumor heterogeneity (5-8). Moreover, detection methods have continued to improve, particularly with the introduction of next-generation sequencing (NGS). NGS can provide more extensive mutation data than the previously used method, amplification refractory mutation system (ARMS), which is widely used to detect hot-spot mutations (5). Hence, the National Comprehensive Cancer Network (NCCN) panel recommends NGS testing for assessment of molecular profiles (9). However, current evidence supporting the recommendation for routine clinical use of ctDNA analysis is conflicting because of the associated levels of false-positive and false-negative results (10).
In this study, we investigated implementation of the liquid biopsy approach in clinical practice, with the aim of facilitating accurate, and correctly interpreted, sequencing results. We also aimed to clarify factors that may influence discordance between genotyping results derived from ctDNA assays and tumor specimens.
In addition, we noted that in a large number of studies on the consistency difference, sequencing of tissue samples occurred at a different time from blood testing and, because of the obstacles of acquiring fresh tissue, previously collected paraffin-embedded tumor specimens were therefore adopted. Further, patients may have received anti-cancer treatments during the period between collection of the two types of specimen, which is expected to generate dynamic alterations in ctDNA (11,12). To this end, we conducted a retrospective analysis of sequencing results from patients who underwent tissue and liquid biopsy simultaneously, which reduces the interference of heterogeneity of temporal treatment, and assessed the consistency between tissue and blood tests. Another key point of this study was to explore indicators that can assist in improving the consistency of ctDNA testing. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-3162).
Methods
Patients and specimen collection
We retrospectively analyzed data from 231 patients with lung cancer who underwent gene sequencing at the Department of Biotherapy of Tianjin Medical University Cancer Hospital from February 2016 to April 2019. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of Tianjin Medical University Cancer Hospital (No. E2016055). Individual consent for this retrospective analysis was waived. The inclusion criteria for this study were as follows: (I) the sequencing panel included the nine driver genes; EGFR, ALK, KRAS, ROS1, RET, MET, PIK3CA, HER-2, and BRAF (9). (II) Tumor DNA and plasma DNA from the patient were simultaneously analyzed; as the turnaround time for testing tumor DNA was longer than that for ctDNA, plasma genotyping was conducted first, resulting in a difference in the times at which tissue and plasma sequencing were conducted. Therefore, “simultaneous” was defined as an interval of <14 days between tissue and blood sampling. (III) Tissue samples used for genotyping were from primary or metastasis tumor samples, while data from analysis of pleural effusion, cerebrospinal fluid, pericardial effusion, etc. were excluded. In total, tumor and plasma samples from 27 patients with lung cancer were analyzed.
Tissue and plasma NGS genotyping
DNA was extracted and isolated using the QIAamp DNAFFPE tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA concentrations were measured using the Qubit dsDNA assay (Thermo Fisher Scientific, Waltham, MA). NGS panels consisted of 8 (3 cases), 56 (1 case), 168 (20 cases), and 520 (3 cases) genes. A similar procedure was used for ctDNA extraction from whole blood. Supernatants were centrifuged for 10 min (16,000 g, 4 °C) then 4 to 5 mL of plasma supernatants were transferred to fresh tubes and extracted using the QIAamp Circulating Nucleic Acid kit (Qiagen, Valencia, CA, USA). Quantification of ctDNA was performed using the Qubit 2.0 Fluorometer and the dsDNA HS assay kit (Life Technologies, Carlsbad, CA, USA). A minimum of 50 ng of ctDNA was required for NGS library construction. Median sequencing depth of tumor and plasma NGS samples were 1,921× and 17,270×, respectively.
NGS library preparation
Extracted tumor DNA was sheared using a Covaris M220 instrument (Covaris Inc, Woburn, MA), followed by end-repair, phosphorylation, and adaptor ligation. Fragments of 200–400 bp were selected using Agencourt AMPure beads (Beckman Coulter, Brea, CA), followed by hybridization with capture probe baits, hybrid selection with magnetic beads, and PCR amplification. A bioanalyzer high-sensitivity DNA assay was conducted to assess fragment quality and size, and indexed samples were sequenced on the NextSeq500 sequencer, with pair-end reads.
Sequencing data analysis
Sequencing data in FASTQ format were mapped to the human hg19 reference sequence (Human Genome version 19) using Burrows-Wheeler Aligner 0.7.10. Genome Analysis Toolkit 3.2, MuTect, and VarScan were used for local alignment optimization, variant calling, and annotation, respectively. DNA translocation analysis was conducted using Tophat2 and Factera 1.4.3.
Statistical analysis and evaluation of tumor DNA and ctDNA data concordance
Concordance was defined as at least one identical driver gene (EGFR, ALK, KRAS, ROS1, RET, MET, PIK3CA, HER-2, BRAF) mutation in both tumor DNA and ctDNA from a patient, or no mutation detected in both tumor DNA and ctDNA from the same patient. Statistical analyses were conducted using SPSS for Windows (version 22.0). The sensitivity and specificity of driver gene mutation detection in plasma samples was defined as the proportion of patients positive and negative for known oncogenic driver mutations detected in tissue (considered the gold standard) who were found to have positive and negative results, respectively, on ctDNA NGS. We applied a non-parametric trend test and Fisher’s exact test to evaluate variance in concordance according to clinicopathological features. All P values are two-sided and P<0.05 was considered statistically significant.
Results
Patient characteristics
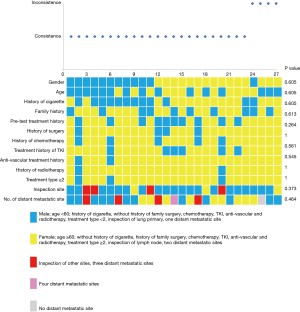
Genotype profiles were collected from 27 patients with lung cancer, consisting of 22 adenocarcinomas, 2 squamous cell carcinomas, 1 sarcomatoid carcinoma, 1 carcinoid, and 1 poorly differentiated carcinoma. As shown in Figure 1, most patients had stage IV tumors (92.6%) and were not receiving therapy (63%). Only 7.4% (n=2) of patients had stage III tumors. Tumor tissue and ctDNA tests were conducted on the same day in 29.6% of patients, and 74.1% of tumor tissue samples were obtained from primary lung tumors. In total, 86 variants were detected in 50 different genes, with 62 and 66 variants detected in tumor and ctDNA sequences, respectively (Figure S1). TP53 and EGFR variants were remarkably frequent in both tumor DNA and ctDNA. Among driver mutations, EGFR exon 19 deletion (19.4% and 14.3%) and exon 21L858R (16.1% and 16.1%), were the most frequently detected in both tumor DNA and ctDNA. The frequencies of single nucleotide variations (SNVs) (n=80 and 76) and copy number variations (CNVs) (n=14 and 13) were not significantly different between tumor DNA and ctDNA samples (P=1.0).
Concordance of driver gene mutations between tDNA and ctDNA
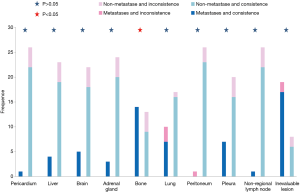
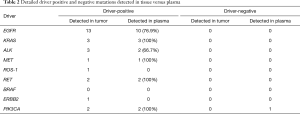
Concordance analysis of synchronous NGS data from the 27 patients with tumor DNA and ctDNA samples showed that overall concordance was 85.2%, sensitivity was 87.0%, specificity was 75%, and the Kappa value was 0.514 (Table 1). Detailed information regarding concordance of driver gene mutations is presented in Table 2. Next, we further explored which factors were associated with inconsistence between tumor DNA and ctDNA data; the results are presented in Figures 1 and 2, and Figure S2. Surprisingly, concordance between ctDNA and tumor DNA was significantly higher in patients with bone metastases relative to those without bone metastases (100% vs. 69.2%, P=0.041) (Figure 2), while no such superior accuracy was detected in patients with metastases at other sites. In addition, other clinical characteristics, including history of therapy, inspection sites, and maximum tumor size showed no significant association with concordance between tumor DNA and ctDNA. Among the reasons for ordering DNA and ctDNA tests, the most common was for initial detection of targetable mutations in 17 patients who had received no previous therapy. Among the remaining patients who received more than first-line treatment, 60% were administered an EGFR-targeted tyrosine kinase inhibitor (TKI), and one patient received a third generation TKI (Osimertinib) in second-line therapy following resistance to gefitinib. Genetic information from the six patients who developed EGFR-TKI resistance are presented in Figure S3. The EGFR mutation, T790M, was identified in one patient (16.7%) from both tumor DNA and ctDNA samples, and an original EGFR-activating mutation (exon 19 deletion) was also detected. Other less common alterations associated with the development of resistance have been reported (13,14), including PTEN deletion (n=1) and TP53 mutation (exon 5 and 6) (n=3). The concordance of detecting alterations in tumor and plasma samples in patients with systemic progression disease was 65.1% (15/23). Among patients who tested negative by either plasma or tumor NGS, the concordance for detecting SNVs was higher than for CNVs (65.5%, 57/87 vs. 20.0%, 4/20, P=0.001).

Full table
Full table
TP53 mutation predicts concordance
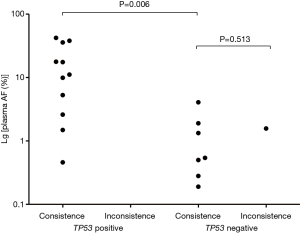
As mentioned above, TP53 mutations were the most common alteration detected. TP53 mutations also commonly co-occurred with changes to driver oncogenes, supported by previous research (15,16). To accurately clarify the co-existence of TP53 and driver mutations, this research exclude patients who test in 8 genes panel not including TP53, for acquiring more gene information. EGFR (58.3%, 7/12) alterations most frequently co-occurred with those of TP53 in ctDNA sequence data, followed by KRAS (16.7%, 2/12), PIK3CA (16.7%, 2/12), and RET (8.3%, 1/12). Surprisingly, our analysis indicated that, where TP53 and driver gene mutations are simultaneously detected in ctDNA, the concordance rate between tumor and ctDNA tests for driver genes was 100% (Table 3). Further, in tests of consistency in samples positive for driver gene mutations, allele frequencies (AFs) of driver alterations in ctDNA from patients with co-mutation of TP53 were significantly higher than those without co-mutation (median: 11.12% vs. 0.54%, P=0.006) (Figure 3). Besides, among these patients with consistent results, a higher number of those with TP53 mutations did not have lung metastasis, relative to those without TP53 mutations (Table S1).

Full table
Discussion
Over the last decade, oncology research has increasingly relied on genomic data, mainly derived from ctDNA samples, shed into blood from primary and metastatic cancers (12). Although tissue biopsy-based NGS is recommended as the primary choice by the NCCN panel, in approximately 14% of cases treated at large academic cancer centers, insufficient tissue is obtained for use in NGS. Furthermore, NGS data from one site may not be representative of the whole tumor because of intra-tumor heterogeneity. Hence, ctDNA analysis has advantages for a proportion of patients. Consequently, NGS of ctDNA is considered complementary to tumor tissue analysis.
Nevertheless, the accuracy and concordance of data from ctDNA, relative to tumor gene sequencing, remains uncertain. In particular, concordance rates are unclear. In this study, the detected concordance rate (85.2%) was superior to those in previously published prospective and retrospective studies assessing the utility of ctDNA (56.6–79%) (2,12,17-20). This discrepancy may be due to the fact that most patients in the present study had advanced lung cancer and hence a greater tumor burden (18); however, a previous study where all enrolled subjects had advanced lung cancer reported a concordance rate of only 56.6% (12), which is substantially lower than our findings. Another potential factor influencing the results is the time interval between the plasma draw date and tissue biopsy, which was longer in the previous study than in this investigation (median: 21 vs. 3 days, interquartile range: 183 vs. 5 days). While there remains no agreement on an appropriate cut-off point for time differences between tissue and blood samples in concordance analysis, many studies have indicated that concordance increases with decreased interval time (2,17,20). Other potential influences, such as tumor evolution or intervening treatment, may also have contributed to the higher rate of concordance found in this study. In order to identify the clinical utility of ctDNA analysis, we further explored the influence of clinical characteristics on concordance. Our findings indicated that patients with bone metastases are found to yield greater accuracy for tissue and plasma in this study. This finding is supported by several previous reports (10,21,22), indicating that greater tumor burden and advanced tumor stage, usually accompanied by bone metastases (11), are promising predictors of higher sensitivity.
This study is the first to report that detection of TP53 mutation in plasma samples is a powerful indicator of reliable ctDNA sequencing of driver genes. This interesting finding relates to lung cancer driver mutations, where mutations of TP53 and other driver genes are the trunk genes in the evolution of lung cancer (7,23). Accordingly, detection of TP53 mutation is a credible marker of the reliability of the results of driver gene analysis. Importantly, in this study there were no patients who were negative for driver gene mutations and for TP53 mutation in plasma. Thus, the predictive power of TP53 mutation could not be determined, and additional investigation to address this issue is warranted. Moreover, we found that AFs of driver alterations in ctDNA were higher in patients with co-mutation of TP53, which will also have reduced the likelihood of false negative findings from ctDNA sequencing.
Conclusions
Overall, our data show that simultaneous sequencing of ctDNA and tumor DNA results in high levels of concordance for driver gene analysis, while the ctDNA approach has the advantage of being minimally invasive. Furthermore, the results of sequencing driver genes in ctDNA are more reliable for lung cancer patients with bone metastasis and those positive for TP53 mutation in ctDNA samples.
Acknowledgments
Funding: The study was supported by a grant from Lian Yun Gang Shi Hui Lan Public Foundation (HL-HS2020-39).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-3162
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-3162
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-3162). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of Tianjin Medical University Cancer Hospital (No. E2016055). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rashdan S, Gerber DE. Immunotherapy for non-small cell lung cancer: from clinical trials to real-world practice. Transl Lung Cancer Res 2019;8:202-7. [Crossref] [PubMed]
- Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016;22:5772-82. [Crossref] [PubMed]
- Spitaleri G, Passaro A, de Marinis F. Ensartinib (X-396) a novel drug for anaplastic lymphoma kinase-positive non-small cell lung cancer patients: we need smart trials to avoid wasting good bullets. Chin Clin Oncol 2019;8:S1. [Crossref] [PubMed]
- Mandel P. Free nucleotides in animal tissues. Prog Nucleic Acid Res Mol Biol 1964;3:299-334. [Crossref] [PubMed]
- Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018;29:1049-55. [Crossref] [PubMed]
- Liang W, Zhao Y, Huang W, et al. Liquid biopsy for early stage lung cancer. J Thorac Dis 2018;10:S876-81. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Butler TM, Spellman PT, Gray J. Circulating-tumor DNA as an early detection and diagnostic tool. Curr Opin Genet Dev 2017;42:14-21. [Crossref] [PubMed]
- National Comprehensive Cancer Network. (NCCN) Clinical Practice Guidelines in Oncology. Small Cell Lung Cancer, Version 1. 2019. Available online: https://www.nccn.org/professionals/physician_gls/pdf/sclc.pdf
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of egfr and kras mutations in advanced lung cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Sabari JK, Offin M, Stephens D, et al. A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J Natl Cancer Inst 2019;111:575-83. [Crossref] [PubMed]
- Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells 2018;7:212. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of tp53 mutations on outcome in egfr-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Smyth EC, Cervantes A. Immunotherapy is not for all comers in chemotherapy-refractory advanced gastric cancer. Better predictive biomarkers are needed. Ann Oncol 2018;29:2027-8. [Crossref] [PubMed]
- Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with kras-mutant non-small cell lung cancer. Clin Cancer Res 2018;24:334-40. [Crossref] [PubMed]
- Schwaederlé MC, Patel SP, Husain H, et al. Utility of genomic assessment of blood-derived circulating tumor DNA (ctdna) in patients with advanced lung adenocarcinoma. Clin Cancer Res 2017;23:5101-11. [Crossref] [PubMed]
- Peng M, Xie Y, Li X, et al. Resectable lung lesions malignancy assessment and cancer detection by ultra-deep sequencing of targeted gene mutations in plasma cell-free DNA. J Med Genet 2019;56:647-53. [Crossref] [PubMed]
- Xu S, Lou F, Wu Y, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett 2016;370:324-31. [Crossref] [PubMed]
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: Results from the actionable genome consortium. Ann Oncol 2019;30:597-603. [Crossref] [PubMed]
- Jia J, Huang B, Zhuang Z, et al. Circulating tumor DNA as prognostic markers for late stage nsclc with bone metastasis. Int J Biol Markers 2018;33:222-30. [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun 2018;9:216. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)