
Narrative review of the novel coronavirus SARS-CoV-2: update on genomic characteristics, transmissions and animal model
Introduction
Coronavirus disease 2019 (COVID-19) which could cause serious respiratory illness has swept the world in a global pandemic. The etiological agent has been identified as a novel coronavirus, now known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is most likely originated from cross-species transmission of zoonotic coronaviruses (1,2). SARS-CoV-2 is highly contagious and COVID-19 represents a spectrum of clinical manifestations. Since the identification of SARS-CoV-2, COVID-19 has triggered enormous human casualties and economic loss. To provide a comprehensive summary to researchers and potential readers, we summarized and discussed the epidemic situation, genomic characteristics, transmission, pathogenicity, animal model, clinical treatment and vaccine to combat the pandemic novel coronavirus.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2084).
Epidemic situation
In the winter of 2019, COVID-19 was firstly identified in China. The SARS-CoV-2 infection was firstly reported to World health Organization (WHO) on 31 December 2019 (3). More than 36 million laboratory-confirmed cases have been reported globally as of middle October 2020, with more than 1 million deaths (4). The pandemic has touched nearly every corner in the world, more than 200 countries and territories were affected by SARS-CoV-2. Multiple public health measures were taken to prevent the import and spread of SARS-CoV-2 in different countries, including widespread mask usage, temperature measuring, social distancing and self-isolation of mild patients, etc. (5,6). Due to the appropriate prevention and control measures in the early outbreak, currently the new cases in China are mainly imported cases. While America and India have become the epicenters of COVID-19 pandemic, thousands of new cases are now being reported every day. As SARS-CoV-2 is a new coronavirus, most of the human population have no pre-existing immunity and are generally susceptible to the infection (7-10). According to multiple studies of laboratory-confirmed cases from China (11-13), fever, fatigue and cough, to a less extent, diarrhea are the most common symptoms. The symptoms of the pediatric infection cases are less severe and nonspecific in contrast to adult patients. Most patients have a good prognosis except the elderly and those with chronic co-morbidities (14,15). Some patients may not develop the typical manifestations (or very mild which were often neglected by patients) as described above (16), so called asymptomatic patients, which makes it more difficult to prevent and control the COVID-19 disease.
Genomic characteristics and origin
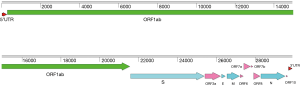
All CoVs share similarities in genome organization and mainly encode four structural proteins including spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins (17). The GC content of SARS-CoV-2 genome is about 38%, with a genome of around 29.8 kb, which also encodes at least six putative accessary proteins, including ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10 that are interspersed between the structural genes according to the reference strain (NCBI accession number: NC_045512.2) of SARS-CoV-2 (Figure 1), though the exact number of functional proteins remains to be established. SARS-CoV-2 showed high nucleotide homology (96.2%) with BatCoV RaTG13 (GenBank: MN996532.1) (18), a bat coronavirus discovered from Rhinolophus affinis in Yunnan Province, China. Genome sequence homology of SARS-CoV-2 with SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) was 76.71% and 33.84%, respectively. Until now more than ten thousand SARS-CoV-2 sequences have been submitted to GISAID (https://www.gisaid.org/). All strains of SARS-CoV-2 genomes are highly conserved and distinguished from other betacoronavirus (Figure 2). SARS-CoV-2 has low genetic diversity due to the presence of proofreading mechanisms. However, some biased mutations may occur and result in the dominant variant due to natural selection, like D614G (A23403G) on spike protein (19-21). Although geographic origin of A23403G cannot be determined, the frequency of this mutation has increased to 74% of all the published sequences, and D614G has been proved to relate to increasing infectivity of SARS-CoV-2 (22,23). Although D614G decreases the affinity for ACE2 by increasing the rate of dissociation, further research revealed that D614G would change the conformation of the S1 domain to increase the probability of existence of open conformations. However, whether COVID-19 severity related to D614G variant has not yet been detected. Most importantly, D614G variants showed equally sensitive with D614 strains to binding SARS-CoV-2 RBD neutralizing monoclonal antibodies (23). As the pandemic continues, more D614G-like mutations would be found, it is necessary to develop SARS-CoV-2 infectious clone (ic) of single-point mutant strains to study the effect of mutant on pathogenicity and transmission.

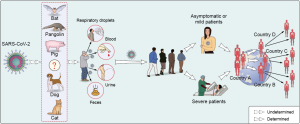
SARS-CoV-2 is believed originated from wild animals (18). It is crucial to discover COVID-19 related animal origins, which will benefit infection prevention and control. A bat coronavirus, RaTG13 has a high nucleotide homology to SARS-CoV-2 (18). Bats are natural reservoir for a number of SARS-related coronaviruses (SARSr-CoVs), Both SARS-CoV and MERS-CoV are zoonotic pathogens which crossed the species barriers to infect humans (1). In detail, SARS-CoV originated from bat (24) and civet cats was the intermediate animals (25,26). MERS-CoV infection in humans was mainly transmitted through close contact with infected dromedary camels (27), while the most recent ancestor analysis speculated that MERS-CoV might have jumped from bats to camels (28). Whether SARS-CoV-2 also originated from bats needs more evidences and further investigation. Another critical question is to identify intermediate hosts that directly transmit SARS-CoV-2 to human. Some of the reported cases identified in December 2019 in Wuhan city were related to Hua-nan seafood market, which had housed a number of wild animals (29). Sellers, consumers, visitors or wildlife housed in the market before the outbreak might be related to SARS-CoV-2 infection and transmission. It is necessary to determine the original source, the transportation route and the destination of the wild animals in the market. However, some COVID-19 patients had no travel history to the seafood market, or even Wuhan City (11,25). According to some most recent reports, the first COVID-19 related cases in France and America might be earlier than previously believed (30). Multiple animal species worldwide were detected to be positive for SARS-CoV-2 RNA by quantitative PCR recently, including cat (31), dog (32), pangolin (33), tiger and lions (34), etc., indicating many animals can be infected with SARS-CoV-2 and have the potential to transmit the virus to humans (35) (Figure 3). However, most of the scientists agree that there is no evidence showing pets spread SARS-CoV-2 to other animal, including human. Global wildlife monitoring needs to be stepped up to reduce the risk of future pandemics. Besides, more important information about potential zoonotic transmission of SARS-CoV-2 infection can be acquired by conducting comparative analysis of ACE2 sequences from different species and predicting their ability of binding SARS-CoV-2 spike protein (36). Comparative and structural analysis of ACE2 in vertebrates showed that a large number of mammals that can potentially be infected by SARS-CoV-2 via their ACE2 proteins, which represents an opportunity for spillover of SARS-CoV-2 from humans to other susceptible animals (36). Further studies are required to identify the exact natural reservoir and intermediary hosts responsible for the COVID-19 pandemic worldwide.
Transmission and pathogenicity
COVID-19 had been announced as a worldwide pandemic by WHO on 11 March 2020, but the transmission mechanism of SARS-CoV-2 has yet to be determined. Comparison the transmission routes of SARS-CoV-2 with SARS-CoV and MERS-CoV may provide useful information against the novel CoV. MERS-CoV infection is mainly due to the repeated introductions of MERS-CoV from dromedary camels to human, resulting in limited human to human transmission (37,38). SARS-CoV is predominantly transmitted through close person-to-person contact, via respiratory droplets (39). As to SARS-CoV-2, nucleic acid shedding pattern of patients infected with SARS-CoV-2 appears different from that had been seen in patients infected with SARS-CoV (40). Higher viral loads were detected soon after symptom onset, and more robust SARS-CoV-2 replication was found in upper respiratory tract. Successful isolation of live virus from nasal and throats swabs is an obvious difference between COVID-19 and SARS (41). Such isolations were rarely successful for SARS-CoV (42,43). In addition, SARS-CoV-2 is stable for several hours to days in aerosols and on surfaces of different kind of materials, suggesting that people may acquire the virus through the air and after touching contaminated objects (44-47). Current epidemiological data indicate that human-to-human transmission of COVID-19 occurs in public and family clusters (48-50). Transmission between infected and healthy individuals can occur through close contact or respiratory droplets, so continuous monitoring of close contacts with confirmed COVID-19 patients is essential. Besides, live viruses can be isolated from specimens collected outside respiratory tract, such as feces (51) and urine samples (52), which suggested that the virus may be transmitted by other routes in addition to close contact (53,54) (Figure 3). Similar transmission route was also observed in the Amoy Gardens outbreak of SARS in 2003 (55). SARS-CoV spread to other residents lived in different apartments through the sewage-disposal system, person-to-person contact, and the use of communal facilities such as elevators and staircases.
As for the pathogenicity, SARS-CoV-2, SARS-CoV and MERS-CoV all cause severe pneumonia with different mortality rates (56,57). By the end of the SARS epidemic, more than 8,096 people had been infected, leading to 774 deaths (9.56%) (58). As for MERS outbreak, a total of 2,562 lab-confirmed cases including 881 (mortality rate 34.4%) has been notified by WHO as of October 2020 (56). However, epidemiological studies that relied on serological testing indicated the underestimate of the prevalence of the MERS-CoV infection (59). Meanwhile, T cell response analysis also indicated that zoonotic infection of dromedary-exposed individuals is taking place in Nigeria and the extent of MERS-CoV infections in Africa is underestimated (60). Mortality rate of COVID-19 was estimated to be about 3.4% as of March by WHO (61). However, the existence of asymptomatic and subclinical SARS-CoV-2 infections makes it difficult to define the actual mortality rate from COVID-19 and the reported mortality rate is very different across the globe. For instance, based on actual case and death numbers in New York City, the fatality rate is about 1.4% (62). Human angiotensin converting enzyme II (hACE2), the cell entry receptor of SARS-CoV, was also proved to be the cell entry receptor for SARS-CoV-2. SARS-CoV-2 spike protein showed a comparable or higher affinity to hACE2, which is consistent with the phenomenon that SARS-CoV-2 has stronger transmissibility (63,64). Human single-cell RNA sequencing (scRNA-seq) datasets showed that organs, including lung, liver, stomach, ileum, kidney and colon have high ACE2 expression, indicating these organs are at potential risk to SARS-CoV-2 infection (65). Like SARS-CoV and MERS-CoV, the cause of severe infection in COVID-19 patients is dysregulated inflammatory response, also called cytokine storm, which leads to acute respiratory distress syndrome and multiple organ failure (14). Pulmonary edema with hyaline membrane formation, desquamation of pneumocytes and hyaline membrane formation were found from autopsy findings of a COVID-19 patient, which confirmed the existence of acute respiratory distress syndrome (66). The positive PCR result of feces and blood sample supported the notion that SARS-CoV-2 infection could be systemic (53). Severe patients may develop dyspnea and hypoxemia one week after the onset of the disease, and some severe cases rapidly progressed to acute respiratory distress syndrome, septic shock, refractory metabolic acidosis, coagulation dysfunction and multi-organ failure (13). Compared with mild patients, patients with severe COVID-19 tended to have a higher viral load and a longer virus-shedding period (67). Severe and critical cases had higher IgM levels than mild cases, whereas the IgG level in critical cases was lower than those in both mild and severe cases (68). Besides, numerous asymptomatic patients were also detected (69,70). These patients were positive for viral RNA detection without any typical COVID-19 manifestations, which may be responsible for the complicated transmission routes of SARS-CoV-2.
Lab diagnostic
As to lab diagnostic, the shared genome by China CDC enables other countries to rapidly diagnose patients by real-time RT-PCR (71-73), WHO also issued guidance on how to detect and treat patients infected with SARS-CoV-2 (74). SARS-CoV-2 virus RNA was mainly detected in respiratory samples, including throat swabs, nasal swabs and sputum. Viral nucleic acid test can remain positive for a long time in feces samples from some severely ill patients. Moreover, severe patients show a prolonged viral shedding than mild patients (75). Multiple rapidly IgA, IgG and IgM antibody detection methods were also established, including lateral flow immunoassay (76), chemiluminescence immunoassay (77) and luciferase immunoprecipitation assay systems (LIPS) (78). Early detection of SARS-CoV-2 antibodies in COVID-19 patients performed as a serologic marker of infection (79) and anti-SARS-CoV-2-NP IgG level correlated with virus neutralization titer (80) which could guide and improve treatments. Mild patients have lower IgM responses against SARS-CoV-2, which indicated lower viral loads, so using IgM detection for sole diagnosis of COVID-19 in patients with mild disease might result in false negative. Meanwhile, viral specific IgG in sputum and urine from severe patients presented positive, so detection of antibody responses in urine and other body fluids could be used to as a marker to determine disease severity (75). CRISPR-Cas12-based nucleotide acid detection assay provides a visual and faster alternative method than real-time RT-PCR assay in clinical departments (81). Detectable SARS-CoV-2 viral RNA in blood is a strong indicator for the progress of severe COVID-19 (82). The ongoing research advances in complementary technologies paved the way to more reliable, accurate and high throughput detection results.
Animal model
Animal models mimicking the clinical symptoms and pathological changes of human SARS-CoV-2 infection were essential for antiviral drug screening, vaccine development and viral pathogenesis research. Animal models used for SARS-CoV and MERS-CoV infection are variable. As for MERS, several generations of mouse and other animal models were developed, including mice transduced with adenoviral vectors expressing hDPP4 (human dipeptidyl peptidase 4, the receptor of MERS-CoV spike protein), hDPP4 transgenic mice (83), hDPP4 knockin mice (84) and nonhuman primate model of common marmoset (85) and rhesus macaque (86). Ad-hDPP4 transduction system is useful for the rapid generation of a murine model in 2–3 weeks. MERS-CoV replicates successfully in mouse lungs and causes modest lung damage. However, the level of hDPP4 expression in different epithelial cells is hard to control (87). hACE2 and hDPP4 transgenic mouse model had been developed for SARS-CoV and MERS-CoV infections. However, the systemic expression of hACE2 or hDPP4 may lead to the atypical symptoms and brain infections of SARS-CoV and MERS-CoV (83,84). Human receptor hDPP4 knockin mouse model with mouse adapted virus is the optimal animal model for MERS-CoV (84), although it needs long period of time to breed and backcross these mice. SARS-CoV-2 fails to invade and replicate in traditional mouse model due to the structural differences in mouse ACE2 (mACE2) compared with human ACE2 (hACE2). Several strategies have been developed to overcome this receptor incompatibility, like generating transgenic mice bearing hACE2 receptor (88-90), developing hACE2 mouse model transduced with adenovirus 5 (Ad5), adeno-associated virus (AAV), or Venezuelan equine encephalitis replicon particles (VRP) expressing hACE2 (91-93), and developing mouse adapted SARS-CoV-2 strain by serial passages in the respiratory tract of mice (94,95). Except for the mouse models mentioned above, others animal models of COVID-19 are still under development, including ferrets (96), Syrian hamsters (97), and macaques (98-100). Multiple animal models are urgently needed to reproduce the clinical disease and pathological changes observed in COVID-19 patients, as well as to provide useful tools for studying SARS-CoV-2 infection, transmission, and screening antiviral drug and developing vaccines.
Clinical treatment and vaccine
No licensed antiviral drugs for COVID-19 treatment are currently available. Repurposed drugs with anti-SARS-CoV-2 potential are urgently needed. Some drugs like chloroquine (101), arbidol (102), remdesivir, traditional Chinese medicine (103) which once were used for treatment against malaria, Influenza virus or Ebola virus, are required to be validated in clinical trials (Table 1). Convalescent plasma from patients recovered from COVID-19 could also be useful for emergency treatment of severe and critically ill patients (104,105). Further, human monoclonal antibody isolated from memory B cells of COVID-19 convalescent patients would be another potential treatment, which were developed for MERS (106) and influenza (107) patients. Several SARS-CoV-2 neutralizing antibodies were recently generated, although validations in animal models and in clinical trials were still needed to ensure the safety and efficacy of these medicines and to avoid adverse effect in humans (108,109) (Table 1).
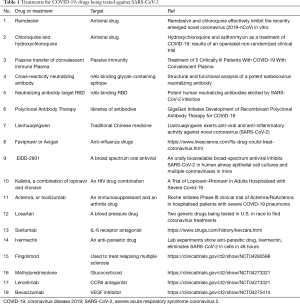
Full table
To fast respond to the COVID-19 pandemic, a safe and effective vaccine is urgently needed. According to the landscape of COVID-19 candidate vaccines prepared by WHO, there are more than 100 candidate vaccines (110,111) under development using various platforms including: nucleic acid (DNA, RNA), protein subunit, viral-vectored, live attenuated and inactivated vaccines some of which has initiated clinical trials. However, different vaccine strategies have certain advantages and disadvantages, it is necessary to evaluate the quality, safety and efficacy of vaccines. Researchers from China reported the initial findings of the first clinical trial for a potential vaccine candidate against SARS-CoV-2 (112). The Ad5 vectored COVID-19 vaccine expressing the spike glycoprotein was tolerable, immunogenic and could induce virus specific humoral and T-cell response after immunization. Multiple inactivated vaccines also showed good performance in animal and human studies (98,113). Significantly, side effect may be induced after immunization; accurate risk assessment plays a key role in vaccination. Recently, trials of a COVID-19 vaccine being developed by AstraZeneca and Oxford University will resume after being paused due to a reported side effect in a patient in the UK. In addition, determining immune correlates of protection for SARS-CoV-2 is also critical for guiding vaccine development, including B cell response and T cell response, which were both important in the control of coronavirus infections. However, the longitudinal study of the protective effect of different vaccines, as well as the potential ADE effect induced by vaccination required further studies. Neutralizing Ab response after SARS-CoV-2 infection is typical of an acute viral infection with declining nAb titres observed following an initial peak (114), a similar phenomenon of nAb declining was also observed in SARS infection, IgG titre reached a peak on day 60, and remained high until day 180 from when it declined gradually until day 720 (115). Determining the longevity of protective immunity is a key problem. At last, most of CoVs were likely originated from cross-species transmission of zoonotic coronaviruses; the concept of immunizing wild animal reservoirs for the prevention of disease in humans is more challenging and has received limited attention.
Summary and outlook
The outbreak of COVID-19 remains a significant public threat. More studies are needed to respond to the challenge. Continuous and vigilance surveillance for SARS-CoV-2 is crucial for epidemic control. Several questions needed to be addressed are listed below.
- Original source and intermediary hosts of SARS-CoV-2 is still unknown. Intensive investigations to identify possible animal reservoir are needed.
- There is an urgent need to develop standard guidelines to treat SARS-CoV-2 infected patients.
- Developing effective and safe vaccines and therapeutics are urgently needed to prevent human infection by SARS-CoV-2.
- Establishment of effective animal model is essential, which could address a variety of scientific questions, including pathogenesis and development of vaccines and therapeutics.
- The understanding of cross-reactivity between SARS-CoV-2 and the other human coronaviruses and the role of pre-existing cross immunity in COVID-19 patients will be essential for patients’ treatment and vaccine design.
Our understanding of COVID-19 is still limited. Many research gaps of SARS-CoV-2 remain to be addressed. The integrated surveillance, response systems and research will facilitate the elimination of the infectious disease. Comprehensive understanding of SARS-CoV-2 could allay public fears and benefit pandemic prevention and control.
Acknowledgments
Funding: This study was funded by grants from The National Key Research and Development Program of China (2018YFC1200100) (Jincun Z), National Science and Technology Major Project (2018ZX10301403) (Jincun Z), the emergency grants for prevention and control of SARS-CoV-2 of Ministry of Science and Technology (2020YFC0841400) and Guangdong province (2020B111108001, 2018B020207013, 2020B111112003), the Guangdong Province Basic and Applied Basic Research Fund (2020A1515010911), National Key Technology R&D Program (2018YFC1311900), Guangdong Science and Technology Foundation (2019B030316028), State Key Laboratory of Respiratory Disease (SKLRD-QN-201715, SKLRD-QN-201912 and SKLRD-Z-202007).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2084
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2084). Prof. NZ serves as an unpaid Editor-in-Chief of Journal of Thoracic Disease. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 2015;23:468-78. [Crossref] [PubMed]
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565-74. [Crossref] [PubMed]
- Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 2013;368:2277-85. [Crossref] [PubMed]
- Gao R, Cao B, Hu Y, et al. Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus. N Engl J Med 2013;368:1888-97. [Crossref] [PubMed]
- Xiao Y, Torok ME. Taking the right measures to control COVID-19. Lancet Infect Dis 2020;20:523-4. [Crossref] [PubMed]
- Jin Y, Yang H, Ji W, et al. Virology, Epidemiology, Pathogenesis, and Control of COVID-19. Viruses 2020;12:372. [Crossref] [PubMed]
- Lee CY, Lin RTP, Renia L, et al. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front Immunol 2020;11:879. [Crossref] [PubMed]
- Ismail AA. Serological tests for COVID-19 antibodies: Limitations must be recognized. Ann Clin Biochem 2020;57:274-6. [Crossref] [PubMed]
- Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. mBio 2020;11:e00722-20. [Crossref] [PubMed]
- Zeberg H, Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 2020;587:610-2. [Crossref] [PubMed]
- Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382:1708-20. [Crossref] [PubMed]
- Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054-62. [Crossref] [PubMed]
- Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020;323:1239-42. [Crossref] [PubMed]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Song TZ, Zheng HY, Han JB, et al. Delayed severe cytokine storm and immune cell infiltration in SARS-CoV-2-infected aged Chinese rhesus macaques. Zool Res 2020;41:503-16. [Crossref] [PubMed]
- Han C, Duan C, Zhang S, et al. Digestive Symptoms in COVID-19 Patients With Mild Disease Severity: Clinical Presentation, Stool Viral RNA Testing, and Outcomes. Am J Gastroenterol 2020;115:916-23. [Crossref] [PubMed]
- Su S, Wong G, Shi W, et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol 2016;24:490-502. [Crossref] [PubMed]
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270-3. [Crossref] [PubMed]
- Fauver JR, Petrone ME, Hodcroft EB, et al. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell 2020;181:990-6.e5. [Crossref] [PubMed]
- Korber B, Fischer WM, Gnanakaran S, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020;182:812-27.e19. [Crossref] [PubMed]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol 2020;81:104260. [Crossref] [PubMed]
- Li Q, Wu J, Nie J, et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020;182:1284-94.e9. [Crossref] [PubMed]
- Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020;183:739-51.e8. [Crossref] [PubMed]
- Hu B, Zeng LP, Yang XL, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. Plos Pathogens 2017;13:e1006698. [Crossref] [PubMed]
- Ralph R, Lew J, Zeng T, et al. 2019-nCoV (Wuhan virus), a novel Coronavirus: human-to-human transmission, travel-related cases, and vaccine readiness. J Infect Dev Ctries 2020;14:3-17. [Crossref] [PubMed]
- Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005;310:676-9. [Crossref] [PubMed]
- Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 2013;13:752-61. [Crossref] [PubMed]
- Corman VM, Ithete NL, Richards LR, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 2014;88:11297-303. [Crossref] [PubMed]
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727-33. [Crossref] [PubMed]
- Available online: https://52proxy.xyz/index.php?q=https%3A%2F%2Fwww.bbc.com%2Fnews%2Fworld-europe-52526554. BBC News, 2020.
- Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020;368:1016-20. [Crossref] [PubMed]
- Sit THC, Brackman CJ, Ip SM, et al. Infection of dogs with SARS-CoV-2. Nature 2020;586:776-8. [Crossref] [PubMed]
- Lam TT, Jia N, Zhang YW, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020;583:282-5. [Crossref] [PubMed]
- Update On the Bronx Zoo on Tiger Which Tested Positive for COVID-19: Nadia’s Condition Improving - Along with the Condition of the Other Tigers and Lions Which Had Similar Symptoms, 2020. WCSNewsroom website.
- Luan J, Lu Y, Jin X, et al. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun 2020;526:165-9. [Crossref] [PubMed]
- Damas J, Hughes G, Keough K, et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A 2020;117:22311-22. [Crossref] [PubMed]
- Reusken CB, Haagmans BL, Müller MA, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 2013;13:859-66. [Crossref] [PubMed]
- Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015;386:995-1007. [Crossref] [PubMed]
- Vijay R, Perlman S. Middle East respiratory syndrome and severe acute respiratory syndrome. Curr Opin Virol 2016;16:70-6. [Crossref] [PubMed]
- Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med 2020;382:1177-9. [Crossref] [PubMed]
- Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465-9. [Crossref] [PubMed]
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-66. [Crossref] [PubMed]
- Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-76. [Crossref] [PubMed]
- van Doremalen N, Bushmaker T, Morris DH. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med 2020;382:1564-7. [Crossref] [PubMed]
- Fennelly KP. Particle sizes of infectious aerosols: implications for infection control. Lancet Respir Med 2020;8:914-24. [Crossref] [PubMed]
- Candido DS, Claro IM. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 2020;369:1255-60. [Crossref] [PubMed]
- Zhang R, Li Y. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc Natl Acad Sci U S A 2020;117:14857-63. [Crossref] [PubMed]
- Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 2020;382:1199-207. [Crossref] [PubMed]
- Tong ZD, Tang A, Li KF, et al. Potential Presymptomatic Transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis 2020;26:1052-4. [Crossref] [PubMed]
- Cai J, Sun W, Huang J, et al. Indirect Virus Transmission in Cluster of COVID-19 Cases, Wenzhou, China, 2020. Emerg Infect Dis 2020;26:1343-5. [Crossref] [PubMed]
- Xiao F, Sun J, Xu Y, et al. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg Infect Dis 2020;26:1920-2. [Crossref] [PubMed]
- Sun J, Zhu A, Li H, et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg Microbes Infect 2020;9:991-3. [Crossref] [PubMed]
- Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020;323:1843-4. [Crossref] [PubMed]
- Ong SWX, Tan YK, Chia PY, et al. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020;323:1610-2. [Crossref] [PubMed]
- Yu IT, Li Y, Wong TW, et al. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N Engl J Med 2004;350:1731-9. [Crossref] [PubMed]
- de Wit E, van Doremalen N, Falzarano D, et al. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14:523-34. [Crossref] [PubMed]
- Porcheddu R, Serra C, Kelvin D, et al. Similarity in Case Fatality Rates (CFR) of COVID-19/SARS-COV-2 in Italy and China. J Infect Dev Ctries 2020;14:125-8. [Crossref] [PubMed]
- Roussel Y, Giraud-Gatineau A, Jimeno MT, et al. SARS-CoV-2: fear versus data. Int J Antimicrob Agents 2020;55:105947. [Crossref] [PubMed]
- Alshukairi AN, Zheng J, Zhao J, et al. High Prevalence of MERS-CoV Infection in Camel Workers in Saudi Arabia. mBio 2018;9:e01985-18. [Crossref] [PubMed]
- Mok CK, Zhu A, Zhao J, et al. T-cell responses to MERS coronavirus infection in people with occupational exposure to dromedary camels in Nigeria: an observational cohort study. Lancet Infect Dis 2020. [Crossref] [PubMed]
- WHO Director-General's opening remarks at the media briefing on COVID-19 - 3 March 2020 - World Health Organization, March 3, 2020.
- Leaf-nosed bat. Encyclopædia Britannica: Encyclopædia Britannica Online; 2009.
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-3. [Crossref] [PubMed]
- Walls AC, Park YJ, Tortorici MA, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020;181:281-92.e6. [Crossref] [PubMed]
- Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185-92. [Crossref] [PubMed]
- Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420-2. [Crossref] [PubMed]
- Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020;20:656-7. [Crossref] [PubMed]
- Hou H, Wang T, Zhang B, et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology 2020;9:e01136. [Crossref] [PubMed]
- Ye F, Xu S, Rong Z, et al. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis 2020;94:133-8. [Crossref] [PubMed]
- Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 2020;63:706-11. [Crossref] [PubMed]
- Waggoner JJ, Stittleburg V, Pond R, et al. Triplex Real-Time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis 2020;26:1633-5. [Crossref] [PubMed]
- LeBlanc JJ, Gubbay JB, Li Y, et al. Real-time PCR-based SARS-CoV-2 detection in Canadian Laboratories. J Clin Virol 2020;128:104433. [Crossref] [PubMed]
- Yip CC, Ho CC, Chan JF. Development of a Novel, Genome Subtraction-Derived, SARS-CoV-2-Specific COVID-19-nsp2 Real-Time RT-PCR Assay and Its Evaluation Using Clinical Specimens. Int J Mol Sci 2020;21:2574. [Crossref] [PubMed]
- WHO. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. Available online: https://www.who.int/publications/i/item/10665-331501
- Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest 2020;130:5235-44. [Crossref] [PubMed]
- Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 2020;92:1518-24. [Crossref] [PubMed]
- Padoan A, Cosma C, Sciacovelli L, et al. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med 2020;58:1081-8. [Crossref] [PubMed]
- Burbelo PD, Riedo FX, Morishima C, et al. Detection of Nucleocapsid Antibody to SARS-CoV-2 is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. medRxiv 2020. doi: 10.1101/2020.04.20.20071423. [Crossref]
- Rongqing Z, Li M, Song H, et al. Early Detection of Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies as a Serologic Marker of Infection in Patients With Coronavirus Disease 2019. Clin Infect Dis 2020;71:2066-72. [Crossref] [PubMed]
- To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020;20:565-74. [Crossref] [PubMed]
- Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 2020;38:870-4. [Crossref] [PubMed]
- Chen W, Lan Y, Yuan X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect 2020;9:469-73. [Crossref] [PubMed]
- Zhao G, Jiang Y, Qiu H, et al. Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with Middle East respiratory syndrome-coronavirus. PLoS One 2015;10:e0145561. [Crossref] [PubMed]
- Li K, Wohlford-Lenane CL, Channappanavar R, et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 2017;114:E3119-28. [Crossref] [PubMed]
- Chan JF, Yao Y, Yeung ML, et al. Treatment With Lopinavir/Ritonavir or Interferon-β1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J Infect Dis 2015;212:1904-13. [Crossref] [PubMed]
- de Wit E, Feldmann F, Cronin J, et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A 2020;117:6771-6. [Crossref] [PubMed]
- Zhao J, Li K, Wohlford-Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 2014;111:4970-5. [Crossref] [PubMed]
- Sun SH, Chen Q, Gu HJ, et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 2020;28:124-33.e4. [Crossref] [PubMed]
- Bao L, Deng W, Huang B, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020;583:830-3. [Crossref] [PubMed]
- Jiang RD, Liu MQ, Chen Y, et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell 2020;182:50-8.e8. [Crossref] [PubMed]
- Israelow B, Song E, Mao T, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 2020;217:e20201241. [Crossref] [PubMed]
- Sun J, Zhuang Z, Zheng J, et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020;182:734-43.e5. [Crossref] [PubMed]
- Hassan AO, Case JB, Winkler ES, et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020;182:744-53.e4. [Crossref] [PubMed]
- Gu H, Chen Q, Yang G, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020;369:1603-7. [PubMed]
- Dinnon KH 3rd, Leist SR, Schäfer A, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020;586:560-6. [Crossref] [PubMed]
- Kim YI, Kim SG, Kim SM, et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020;27:704-9.e2. [Crossref] [PubMed]
- Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020;369:956-63. [Crossref] [PubMed]
- Gao Q, Bao L, Mao H, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020;369:77-81. [Crossref] [PubMed]
- Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020;585:268-72. [Crossref] [PubMed]
- Chandrashekar A, Liu J. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020;369:812-7. [Crossref] [PubMed]
- Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30:269-71. [Crossref] [PubMed]
- Zhu Z, Lu Z, Xu T, et al. Arbidol Monotherapy is Superior to Lopinavir/ritonavir in Treating COVID-19. J Infect 2020;81:e21-3. [Crossref] [PubMed]
- Runfeng L, Yunlong H, Jicheng H, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res 2020;156:104761. [Crossref] [PubMed]
- Chen L, Xiong J, Bao L, et al. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis 2020;20:398-400. [Crossref] [PubMed]
- Shen C, Wang Z, Zhao F, et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA 2020;323:1582-9. [Crossref] [PubMed]
- Corti D, Zhao J, Pedotti M, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci U S A 2015;112:10473-8. [Crossref] [PubMed]
- Henry Dunand CJ, Leon PE, Huang M, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 2016;19:800-13. [Crossref] [PubMed]
- Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020;584:120-4. [Crossref] [PubMed]
- Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020;584:115-9. [Crossref] [PubMed]
- Amanat F, Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020;52:583-9. [Crossref] [PubMed]
- Karpas A, Dremucheva A, Czepulkowski BH. A human myeloma cell line suitable for the generation of human monoclonal antibodies. Proc Natl Acad Sci U S A 2001;98:1799-804. [Crossref] [PubMed]
- Zhu FC, Li YH, Guan XH, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020;395:1845-54. [Crossref] [PubMed]
- Wang H, Zhang Y, Huang B, et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020;182:713-21.e9. [Crossref] [PubMed]
- Seow J, Graham C, Merrick B, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv 2020. doi: https://doi.org/. [Crossref]
- Mo H, Zeng G, Ren X, et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 2006;11:49-53. [Crossref] [PubMed]


