
SAMHD1 can suppress lung adenocarcinoma progression through the negative regulation of STING
Introduction
Despite the identification of aberrant signaling pathways in non-small cell lung cancer (NSCLC), the pathogenetic mechanisms remain to be elucidated. The identification of essential driver mutations associated with the carcinogenesis of NSCLC have radically altered the treatment strategy for this disease (1). New drugs have been developed that specifically target the key mutated proteins, such as epidermal growth factor receptor (EGFR), Kirsten ras oncogene homolog (KRAS), and anaplastic lymphoma kinase (ALK) mutant forms (2-4). However, the overall 5-year survival rate for lung cancer remains at just 15% (5). Therefore, improving our understanding of the pathophysiological processes in tumor cells that promote local aggression and recurrence in NSCLC is important.
The sterile alpha motif (SAM) domain and histidine-aspartate (HD) domain-containing protein 1 (SAMHD1) is a deoxynucleoside triphosphate (dNTP) phosphohydrolase that cleaves dNTPs into deoxynucleosides and inorganic triphosphates, thus depleting the cellular dNTP pool required for cellular DNA polymerase (6-8). Recruitment of SAMHD1 to DNA repair foci also happens in response to DNA damage (9). We previously reported in a small-sample-sized study that SAMHD1 was downregulated at both protein and messenger RNA (mRNA) levels in LAC compared with adjacent healthy tissues. We also found that overexpression of SAMHD1 inhibited the proliferation of lung tumor cells (10). These results revealed the regulation and function of SAMHD1 in lung cancer, which is important for the proliferation of lung tumor cells. However, the mechanism underlying the effect of SAMHD1 and the downstream target genes in LAC remains unclear.
The stimulator of interferon genes (STING) is a 379-amino acid transmembrane-protein located in the endoplasmic reticulum. As a DNA sensor, STING is activated by cyclic-dinucleotides (CDNs) (11,12). Evidence shows that the STING pathway is involved in tumorigenesis, and it has both inhibitory and promoting effects in tumor development (13-15). However, its regulation effect in lung cancer is not apparent and needs to be elucidated.
The objectives of this study were thus to evaluate the suppressive effect of SAMHD1 on proliferation and apoptosis in LAC and explore the downstream targets of SAMHD1. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-1889).
Methods
Participant information collection
A total of 238 patients who underwent surgical resection for primary NSCLC at the Department of Thoracic Surgery, Huadong Hospital Affiliated with Fudan University, between January 2017 and December 2019 were enrolled in this retrospective study. The inclusion criteria were as follows: (I) patients who were diagnosed with primary LAC based on clinical features, radiological examination, and pathological confirmation; (II) age ≥18 years; (III) fresh tumor tissues and paired adjacent tissues were collected during surgical resection, snap-frozen in liquid nitrogen, and stored at –80 °C in the hospital storage room. The exclusion criteria were as follows: (I) insufficient baseline data and follow-up information; (II) neoadjuvant therapy received before surgery; (III) history of other solid tumors, hematologic malignancy, serious infection, or severe hepatic or renal dysfunction. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Signed informed consent was provided by all participants, and the Clinical Research Ethics Committee of Huadong Hospital Affiliated with Fudan University approved the study (2019K104).
Cell culture and lentivirus infection
The NSCLC cell line A549 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). It was inspected by mycoplasma detection, DNA fingerprinting, isozyme detection, and cell vitality detection. All of the cells were cultured in Roswell Park Memorial Institute Medium (RPMI) 1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific), 100 U/mL penicillin G, and 100 streptomycin (GIBCO, Grand Island, NY, USA) in a humidified incubator at 37 °C with 5% CO2. Lentivirus-overexpressing SAMHD1/STING or containing control empty vector was purchased from HanBio (Shanghai, China). The A549 cells were cultured in the six-well plates. After reaching 80% confluence, medium containing lentiviruses and polybrene (8 µg/mL, HanBio) were mixed with cells at a multiplicity of infection (MOI) of 10. After incubation for 24 h, supernatants in the wells were replaced with RPMI 1640 supplemented with 10% FBS, and then cultured for 36 h. The DNA damage response inhibitor (DDRi) was purchased from PeproTech, Inc. (Rocky Hill, NJ, USA).
Quantitative reverse-transcription polymerase chain reaction (RT-qPCR)
The total RNA was isolated using the total RNA kit (Tiangen, Ltd., Haimen, Nantong, Jiangsu, China). For RT-PCR, 1.0 µg of RNA was used with Quantscript RT Kit with gDNase (Tiangen, Ltd.). The potential trace amounts of DNA were removed using gDNase, then complementary DNA was generated. According to the manufacturer’s protocols, quantitative (q)PCR was performed with 500 ng cDNA using 2xTaq PCR MasterMix (Tiangen, Ltd.) and the ABI7500 instrument (Applied Biosystems Instrument, Life, Waltham, MA, USA). The relative expression levels were calculated using the 2ΔΔCt method following normalization against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for SAMHD1. The primers were designed as follows:
SAMHD1-F, 5'-AAAACCAGGTTTCACAACTTCTGC-3'; SAMHD1-R, 5'-TGCGGCATACAAACTCTTTCTGT-3'; STING-F, 5'-CAAGGACCAACTACAACC-3'; STING-R, 5'-TGCCTCTTCTTTAATTG-3'; GAPDH-F, 5'-TGACTTCAACAGCGACACCCA-3'; GAPDH-R, 5'-CACCCTGTTGCTGTAGCCAAA-3';
Western blot analysis
Total proteins were washed with cold phosphate-buffered saline (PBS), extracted in modified radio immunoprecipitation assay (RIPA) buffer, and quantified by the bicinchoninic acid (BCA) method (Beyotime, Beijing, China). Then, 15 µL of proteins were loaded and resolved in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using the Tris-SDS buffer. Proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Burlington, MA, USA) using a wet transfer apparatus (BioRad, Hercules, CA, USA). The membranes were immunoblotted with the following antibodies: anti-SAMHD1 (Abcam, Cambridge, UK), anti-STING (Abcam), and anti-GAPDH antibodies (Abcam). The immunoreactive bands were detected using enhanced chemiluminescence (ECL) Plus reagent (Beyotime). The software ImageJ (https://imagej.nih.gov/ij) was used to quantify the data from three independent experiments.
Cell proliferation and colony formation assays
Cell growth status was analyzed by two methods: the Celigo imaging cytometry system, and 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. The slides were seeded at a density of 104 cells to complete the cell slide for subsequent cytofluorimetric assay. The fluorescence intensity and cells were automatically calculated by the Celigo imaging cytometry system (Nexcelom, Lawrence, MA, USA). We defined cell viability by MTT (Roche, Indianapolis, IN, USA) according to the manufacturer’s instructions (absorbance was measured at 570 nm).
For colony formation assays, cells were plated at a low density (500–1,000 cells/well) onto 6-well plates and observed for 14 days. Colonies were then fixed in 4% paraformaldehyde, stained with 0.5% crystal violet staining solution, and counted.
Migration and invasion assays
Transwell chambers with and without Matrigel (Corning Life Sciences, Corning, NY, USA) were used for cell migration and invasion detection, respectively. Briefly, 104 cells were seeded onto Transwell membrane inserts (Corning Life Sciences) in serum-free medium. Media containing 10% FBS was added to the lower chamber. After incubation for 24 h, the cells in the Transwell upper chamber surfaces were erased, and the cells on the lower surfaces were fixed and stained with crystal violet. The stained cells were observed and calculated in five random fields. For the tumor cell invasion assay, the membrane was pre-coated with 15 µL of Matrigel (BD Biosciences, Bedford, MA, USA), and the subsequent steps were identical to those of the tumor cell migration assay, except cells were incubated for 36 h.
Apoptosis assay
The cells were washed with cold PBS, and then fixed with 75% ethanol overnight at –20 °C. The fixed cells were washed with cold PBS and then stained with Annexin V-APC (Beyotime). After incubation in the dark at room temperature for 15 min, cells were analyzed by FACS Calibur flow cytometer (BD Bioscience, San Jose, CA, USA) in triplicate.
Xenograft tumor formation assay
A total of 15, 6-week-old BALB/c nude mice were obtained from Shanghai Laboratory Animals Center (SLAC) Co., Ltd. (Shanghai, China) and fed in accordance with the regulations and internal biosafety and bioethics guidelines.
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Dunwill Medical Technology (IACUC issue no. 201812006). Experiments were performed under a project license (no. 201812006) granted by the institutional committee board of Shanghai Dunwill Medical Technology, in compliance with Shanghai Dunwill Medical Technology institutional guidelines for the care and use of animals.
The A549 cells were infected with lentivirus for 36 h, and were then subcutaneously injected into the mice at an inoculation dosage of 1×107 cells. The tumor volume (V) was calculated according to the following formula: V = (length × width2)/2; and determined using a digital caliper at 4, 7, 11, 15, 19, and 23 days after implantation. After 3 weeks, the mice were sacrificed, tumor tissue samples were harvested, and their weights were measured. Experiments were performed under a project license (201812006) granted by the institutional committee board of Shanghai Dunwill Medical Technology, in compliance with Shanghai Dunwill Medical Technology institutional guidelines for the care and use of animals.
Statistical analysis
Statistical analysis was performed using SPSS Software 22.0 (IBM Corp., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad, La Jolla, CA, USA). Data are expressed as mean ± SD. Comparisons between the two groups were determined by the t-test, Wilcoxon signed-rank sum test, chi-square test (χ2), and analysis of variance (ANOVA). A value of P<0.05 was considered statistically significant.
Results
The mRNA expression of SAMHD1 was lower in the NSCLC tissues than in the paired adjacent tissue and was associated with advanced disease
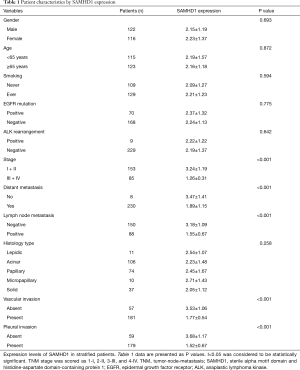
The Wilcoxon signed-rank sum test was used to analyze SAMHD1 expression in tumor and paired adjacent tissues. As shown in Figure 1, SAMHD1 expression was lower in tumor tissues [2.301 (0.686–5.802)] than in paired adjacent tissues [5.679 (3.461–10.783)] (P<0.001). As shown in Table 1, the mRNA level of SAMHD1 was associated with the progression of NSCLC. Furthermore, the expression levels of SAMHD1 were significantly lower in the metastasis-positive patients compared with the metastasis-negative patients (1.89±1.15 vs. 3.47±1.41, P<0.001).
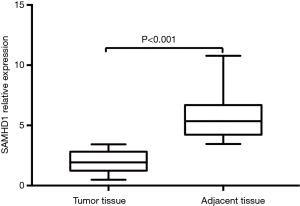
Full table
Overexpression of SAMHD1 inhibited the proliferation, migration, invasion, and expression levels of STING on A549 cells
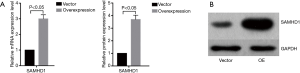
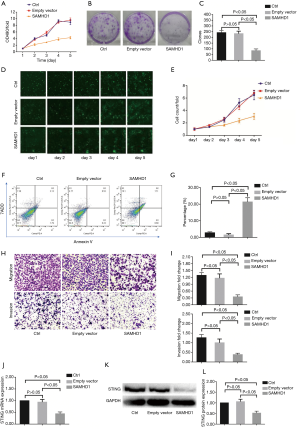
To study the effect of SAMHD1 and STING on A549 cells, the human SAMHD1/STING gene was stably transduced into A549 cells with a SAMHD1/STING expressing lentivirus. Both of their expression levels were determined by RT-qPCR and western blot (Figure 2A,B, P<0.05). The MTT assays indicated that SAMHD1 dramatically inhibited A549 cell growth (Figure 3A, P<0.05). The same results were obtained by colony formation assay (Figure 3B,C, P<0.05) and Celigo cell counter assay (Figure 3D,E, P<0.05). We also analyzed the effect of both SAMHD1 and STING expression on cell apoptosis. Flow cytometry assay revealed that overexpression of SAMHD1 induced cell apoptosis in A549 cells (Figure 3F,G, P<0.05). Thus, our data suggested that SAMHD1 acts as a tumor suppressor in NSCLC cells. For further insights into the response of A549 cells to SAMHD1 expression, we analyzed its effect on migration and invasion behaviors. Transwell assays revealed that SAMHD1 expression suppressed A549 cell migration and invasion (Figure 3H,I, P<0.05). Finally, the mRNA expression level could be inhibited by the overexpression of SAMHD1 (Figure 3J,K,L, P<0.05).
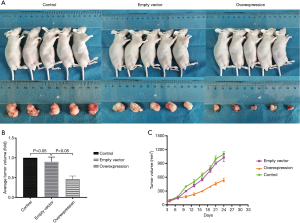
SAMHD1 suppressed tumorigenicity in vivo
We investigated the effect of SAMHD1 on cell growth in vivo. As shown in Figure 4A, tumor growth was significantly slower in SAMHD1-overexpressed mice (n=5) as compared with the control group (n=5) (P<0.05). After 3 weeks, the mice were sacrificed and the tumor tissues were harvested. We found that tumor volumes were lower in the SAMHD1-overexpressed group than in the control group (Figure 4B,C; P<0.05).
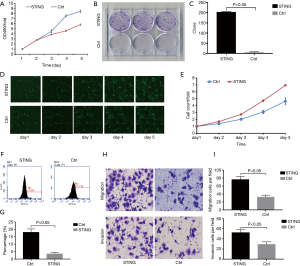
Overexpression of STING weakened the inhibition of SAMHD1
To detect the specific effect of STING for SAMHD1 regulation on A549 cells, the human STING gene was stably transduced into A549 cells with a STING-expressing lentivirus. Both MTT and cell clone assays were measured to detect cellular proliferation. The MTT assays indicated that STING could promote A549 cell growth (Figure 5A; P<0.05). The same results were obtained with colony formation assay (Figure 5B,C; P<0.05) and Celigo cell counter assay (Figure 5D,E; P<0.05). We also analyzed the effect of STING expression on cell apoptosis. Flow cytometry assay showed that overexpression of STING suppressed cell apoptosis in A549 cells (Figure 5F,G; P<0.05). Finally, we analyzed its effect on migration and invasion behaviors. Transwell assays indicated that STING expression could facilitate A549 cell migration and invasion (Figure 5H,I; P<0.05).
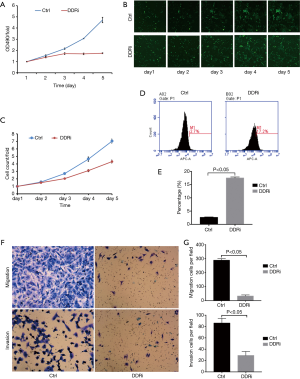
DDRi inhibited migration and invasion of A549 cells
For further verification of the potential regulation of SAMHD1 on the A549 cells and exploration of the definite pathway, DDRi was used in the MTT, Celigo cell counting, apoptosis, and Transwell assays. Both MTT (Figure 6A; P<0.05) and Celigo cell counting assays (Figure 6B,C; P<0.05) showed that DDRi could inhibit the proliferation of A549 cells. Flow cytometry assay indicated that DDRi could promote cell apoptosis in A549 cells (Figure 6D,E; P<0.05). Transwell assays showed that DDRi hindered the A549 cell migration and invasion (Figure 6F,G; P<0.05).
Discussion
Although the improvement of surgical technique and target therapy has been achieved in lung cancer treatment, the 5-year survival rate has been slow to improve. The development of LAC progression involves both genetic changes and tumor environment remodeling, while the deactivation of tumor suppressor genes could induce tumorigenesis and promote the progression of cancer. One report indicated that SAMHD1 could suppress cancer cell proliferation, but its regulatory mechanism is still poorly understood (10).
In this study, we found that a lower expression of SAMHD1 was associated with advanced disease, while the overexpression of SAMHD1 significantly suppressed the apoptosis rate and proliferative abilities of LAC cells. Treatment with the DDRi could inhibit tumor cell proliferation in SAMHD1-overexpressed cells. Overexpression of SAMHD1 could inhibit tumor progression in the xenograft model. The expression of STING was partially suppressed by SAMHD1. Furthermore, the results of rescue experiments indicated that SAMHD1 regulates LAC progression by the repression of STING expression. These results demonstrated that SAMHD1 had a suppressive effect on lung cancer invasion and metastasis, and STING was a potential downstream target of SAMHD1 in lung cancer.
Upregulation of SAMHD1 has been reported to be a predictive factor of poor prognosis in colon cancer, and SAMHD1 was found to affect the chemotherapy response in leukemia (16,17). The results of this study indicated that SAMHD1 could potentially provide information on the mechanisms underlying cancer invasion and metastasis in lung cancer (Table 1), as the involvement of lymph nodes and vascular invasion reflected the invasiveness of tumor cells in a manner partly dependent on the expression level of SAMHD1.
The function of SAMHD1 in tumors remains controversial, as it can reduce not only the efficacy of antimetabolite-based chemotherapies but also has a tumor-suppressive function. SAMHD1 is also potential driver gene in chronic lymphatic leukemia, where it is recurrently mutated in the early stages of tumor development (18,19). Herold et al. reported that SAMHD1 may be a barrier against the therapeutic efficacy of several clinically used nucleoside analogs, and assessment of SAMHD1 levels may allow patient-specific dose adjustments, while targeting SAMHD1 may increase the efficacy of treatment (7,19). In solid tumors, our results indicated that SAMHD1 suppressed the progression of LAC cells in vivo and in vitro.
Tumor progression, which involves metastasis and cell proliferation, can generate cytoplasmic DNA (CDNs), which can be recognized by STING, and then activate the downstream STING pathway (20-23). Recent studies have found that SAMHD1 has an impact on CDNs. Coquel et al. found that SAMHD1 can prevent the accumulation of CDNs because, with the aberrant processing of stalled forks in SAMHD1-depleted human cells, single-stranded DNA fragments are released from stalled forks and accumulate in the cytosol, where they can activate the CDN cyclic 2'3'-GMP-AMP (cGAMP) induced by cGAMP synthase (cGAS), and eventually activate the STING pathway (24). It was found that cGAS can interact with STING through a second messenger, the cGAMP synthesized by cGAS from adenosine triphosphate (ATP) and guanosine triphosphate (GTP), which then directly binds and activates STING (25,26). It is tempting to speculate that SAMHD1 decreases cytosolic DNA and blocks the STING pathway in lung cancer cells; cGAS might play an important role in this progression.
The initial discovery of SAMHD1 was as a dNTP hydrolase that depletes intracellular dNTP pools in non-cycling cells as an antiviral action. Clifford et al. found that SAMHD1 was mutated in chronic lymphocytic leukemia (CLL) and involved in the DNA damage response (16). Daddacha et al. showed that SAMHD1 is recruited to DNA repair foci in response to DNA damage and plays a key role in DNA double-strand break (DSB) repair by promoting the resection of DNA ends (9). In this study, we found that treatment with DDRi could significantly suppress the apoptosis rate and the proliferative abilities in SAMHD1-overexpressing A549 cell lines compared with the control group. Combined with our previous results which that overexpression of SAMDH1 could reduce dNTP level, the present findings suggest that SAMHD1 regulation of LAC progression does not only rely on dNTP hydrolase but also involves DNA damage response.
This study had several limitations. First, we confirmed that SAMHD1 could negatively mediate STING in lung cancer, but the exact mechanism of this action remains unknown. Second, the SAMHD1-STING axis regulates LAC progression in vivo and in vitro, but their downstream target is still not clear. We will investigate these questions in further experiments.
Conclusions
Our study demonstrated that SAMHD1 had a suppressive effect on invasion and metastasis in lung cancer, and STING was identified as a potential downstream target of SAMHD1.
Acknowledgments
The authors would also like to thank the English language editing service of AME Publishing.
Funding: This work was supported by the National Youth Science Foundation (81702252), the Health and Planning Commission Youth Program of Shanghai (N20164Y0160), the ShengKang Developing Program of Shanghai (SHDC12019X11), the Health and Planning Commission Program of Shanghai (201840188), and the Shanghai Science Foundation (18ZR1413000).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-1889
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1889
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1889). The authors have no conflicts of interest to declare.
Ethnical Statement: The authors are accountable for all aspects of this work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). Signed informed consent was provided by all participants, and the Clinical Research Ethics Committee of Huadong Hospital Affiliated with Fudan University approved the study (2019K104). All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Dunwill Medical Technology (IACUC issue no. 201812006). Experiments were performed under a project license (no. 201812006) granted by the institutional committee board of Shanghai Dunwill Medical Technology, in compliance with Shanghai Dunwill Medical Technology institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Reungwetwattana T, Weroha SJ, Molina JR. Oncogenic pathways, molecularly targeted therapies, and highlighted clinical trials in non-small-cell lung cancer (NSCLC). Clin Lung Cancer 2012;13:252-66. [Crossref] [PubMed]
- Chapman AM, Sun KY, Ruestow P, et al. Lung cancer mutation profile of EGFR, ALK, and KRAS: meta-analysis and comparison of never and ever smokers. Lung Cancer 2016;102:122-34. [Crossref] [PubMed]
- Ulivi P, Chiadini E, Dazzi C, et al. Nonsquamous, non-small-cell lung cancer patients who carry a double mutation of EGFR, EML4-ALK or KRAS: frequency, clinical-pathological characteristics, and response to therapy. Clin Lung Cancer 2016;17:384-90. [Crossref] [PubMed]
- Li W, Qu J, Xu Z. Clinical features and mutation status of EGFR, KRAS, BRAF, EML4-ALK and ROS1 between surgical resection samples and non surgical resection samples in lung cancer. J Thorac Dis 2015;7:875-80. [PubMed]
- Rosell R, Neninger E, Nicolson M, et al. Pathway targeted immunotherapy: rationale and evidence of durable clinical responses with a novel, EGF-directed agent for advanced NSCLC. J Thorac Oncol 2016;11:1954-61. [Crossref] [PubMed]
- Li M, Zhang D, Zhu M, et al. Roles of SAMHD1 in antiviral defense, autoimmunity and cancer. Rev Med Virol 2017. [Crossref] [PubMed]
- Herold N, Rudd SG, Sanjiv K, et al. SAMHD1 protects cancer cells from various nucleoside-based antimetabolites. Cell Cycle 2017;16:1029-38. [Crossref] [PubMed]
- Rentoft M, Lindell K, Tran P, et al. Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc Natl Acad Sci U S A 2016;113:4723-8. [Crossref] [PubMed]
- Daddacha W, Koyen AE, Bastien AJ, et al. SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep 2017;20:1921-35. [Crossref] [PubMed]
- Wang JL, Lu FZ, Shen XY, et al. SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochem Biophys Res Commun 2014;455:229-33. [Crossref] [PubMed]
- Takaoka A, Wang Z, Choi MK, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007;448:501-5. [Crossref] [PubMed]
- Lemos H, Huang L, McGaha T, et al. STING, nanoparticles, autoimmune disease and cancer: a novel paradigm for immunotherapy? Expert Rev Clin Immunol 2015;11:155-65. [Crossref] [PubMed]
- Corrales L, Gajewski TF. Molecular pathways: targeting the stimulator of interferon genes (STING) in the immunotherapy of cancer. Clin Cancer Res 2015;21:4774-9. [Crossref] [PubMed]
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016;17:1142-9. [Crossref] [PubMed]
- Corrales L, McWhirter SM, Dubensky TW Jr, et al. The host STING pathway at the interface of cancer and immunity. J Clin Invest 2016;126:2404-11. [Crossref] [PubMed]
- Clifford R, Louis T, Robbe P, et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 2014;123:1021-31. [Crossref] [PubMed]
- Yang CA, Huang HY, Chang YS, et al. DNA-sensing and nuclease gene expressions as markers for colorectal cancer progression. Oncology 2017;92:115-24. [Crossref] [PubMed]
- Sze A, Belgnaoui SM, Olagnier D, et al. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 2013;14:422-34. [Crossref] [PubMed]
- Rudd SG, Schaller T, Herold N. SAMHD1 is a barrier to antimetabolite-based cancer therapies. Mol Cell Oncol 2017;4:e1287554. [Crossref] [PubMed]
- Maelfait J, Bridgeman A, Benlahrech A, et al. Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep 2016;16:1492-501. [Crossref] [PubMed]
- Mackenzie KJ, Carroll P, Martin CA, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017;548:461-5. [Crossref] [PubMed]
- Harding SM, Benci JL, Irianto J, et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017;548:466-70. [Crossref] [PubMed]
- Burdette DL, Monroe KM, Sotelo-Troha K, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011;478:515-8. [Crossref] [PubMed]
- Coquel F, Silva MJ, Técher H, et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 2018;557:57-61. [Crossref] [PubMed]
- Wu J, Sun L, Chen X, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013;339:826-30. [Crossref] [PubMed]
- Ablasser A, Goldeck M, Cavlar T, et al. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature 2013;498:380-4. [Crossref] [PubMed]
(English Language Editors: J. Jones and J. Gray)


