
Mutational signatures in squamous cell carcinoma of the lung
Introduction
Lung cancer is a leading cause of cancer-related death in the world, with 1.76 million cases worldwide and 74,300 cases in Japan (1,2). Although recent advances in molecular targeting therapy and genetic profiling have improved the overall survival in advanced lung cancer patients, these treatments rarely cure lung cancer. In addition, effective molecular targets for therapy for lung squamous cell carcinoma have not been identified. In our previous study, we were unable to identify genetic alterations that could serve as therapeutic targets (3).
Recent advances in immune checkpoint inhibitors have shown promise for lung cancer patients without driver mutations. The programmed cell death-1 (PD-1)/programmed death-ligand 1 (PD-L1) pathway enables tumors to escape from immunosurveillance even if the tumor expresses neo-antigens (4). These neo-antigens usually arise from passenger mutations, and therefore tumor mutation burden (TMB), along with tumor PD-L1 expression, has drawn attention as a biomarker for anti-PD-1/PD-L1 therapy (5). Several kinds of mutagens, such as cigarette smoke, age, and insufficient DNA damage response have been reported to have an influence on TMB and PD-L1 expression (6). However, our previous study could not clarify the factors related with tumor PD-L1 expression and/or TMB in lung squamous cell carcinoma.
The mutational signature is a tool to identify the mutagens that contribute to the mutational spectrum of a tumor by investigating the pattern of DNA changes. Thirty distinct signatures have been identified, and signature 4 (SI4) is related to tobacco smoking with transcriptional strand bias for C>A (and also T>A mutations). SI4 is very similar to the mutational signature induced by exposing cells to benzo(a)pyrene in vitro (7).
Here, we examined the mutagens that may be related to PD-L1 expression and TMB in lung squamous cell carcinoma by performing mutational signature analysis. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2602).
Methods
Patients
Sixty-seven patients who underwent surgical resection of primary lung squamous cell carcinoma between December 2010 and January 2016 at Kyushu University Hospital as well as patients who underwent the same surgery from May 2009 to January 2016 at Niigata University Hospital were included in the study. Patient selection and library preparation for comprehensive genomic sequencing were previously described (3). Clinicopathological features, including age at surgery, smoking history, and pathologic TNM stage (7th edition), were examined (8). This study was approved by the Institutional Review Boards of both the Department of Surgery and Science, Graduate School of Medical Sciences, Kyushu University Hospital (IRB number: 674-01) and the Division of Thoracic and Cardiovascular Surgery, Niigata University Hospital (IRB number: 774). Written informed consent was obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Mutational signature analysis
Comprehensive genomic sequencing was performed using CANCERPLEX v3 (KEW Inc., Cambridge, MA, USA), which is a panel consisting of 415 genes covering 1.66 million base pairs. Variant annotation and filtering were performed as described previously (9).
Mutational signature analysis was performed using MutationalPatterns, R version 3.5.1 (10). A mutation matrix of the 96 mutation type classification was made from each sample’s Variant Call Format (VCF) data, based on the six substitution subtypes and by incorporating information on the bases immediately 5' and 3' to each mutated base. Alexandrov et al. proposed 30 distinct signature patterns from curated data generated by The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) using non-negative matrix factorization (7). SI7, SI4, SI6/15, SI2/13, and SI5 were the main signatures in non-small cell lung cancer in an analysis of 1,144 samples (6). These signatures are related to exogenous mutagens, with SI7 related to ultraviolet (UV), SI4 to smoking, SI6 and S15 to mismatch repair (MMR) defect, SI2 and S13 related to apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC), and SI5 to aging or “clock-like” (7). We used these seven signatures instead of deciphering them from our own data because of the small number of total mutation counts in our datasets.
Immunohistochemistry
Immunohistochemistry for PD-L1 was performed using formalin-fixed paraffin embedded tissue sections, as described in our previous study (11). The primary antibody was an anti-human PD-L1 rabbit monoclonal antibody (clone SP142, dilution 1:100; Spring Bioscience, Ventana, Tucson, AZ, USA). The PD-L1 tumor proportion score (TPS) was estimated as the percentage of total carcinoma cells positive for PD-L1 expression in whole sections, and a cut-off value of 5% was adopted.
Statistical analysis
Statistical significance between groups was determined using the Mann-Whitney U test and Spearman’s rank correlation test. P values of less than 0.05 were considered to indicate statistical significance.
Results
Mutation signature reconstruction using targeted sequencing data
Mutational signature analysis is usually performed using datasets from whole genome sequences (3 billion base pairs) or whole exome sequences (30 million base pairs). We thus performed a preliminary analysis using publicly available TCGA data to determine whether the mutational signature analysis could be performed on a mutation matrix based on a CANCERPLEX mutation dataset (covering 1.66 million base pairs of genomic DNA). As an approximated hypothetical Mutation Annotation Format (MAF) data, mutation data of the 415 genes used for CANCERPLEX were extracted from a MAF file of whole exosome sequencing data from 487 lung squamous cell cancer patients (12). Mutational signatures were then analyzed using Maftools, R version 0.9.30 (13) from the virtual MAF data and from the original MAF data. Although the proportion of contribution for each signature was different, three estimated signatures, SI4, SI7, and SI13, were the same, suggesting that the mutational data of 415 genes is enough to estimate the types of mutational signatures (Figure S1).
The relationship between TMB and smoking or PD-L1 expression
The characteristics of the 67 patients were previously reported (3), for example: 60 (89.6%) patients were male, all patients were smokers with median pack-year index (PYI) of 50 (range: 7–150), and the median age of all patients was 70 (range: 47–84) years (3). Mutational signatures were calculated from VCF data from each sample, and mutation counts were then reconstructed. The reconstructed mutation counts ranged from 13.3 to 98.7, with a median count of 30.1 (Figure 1). Fifty-six patients (83.6%) had more than four signatures, with SI6 as the most frequently observed (64 cases, 95.5%). Other signatures were observed as follows: SI2 in 26 cases (38.8%), SI4 in 45 (67.2%), SI5 in 59 (88.1%), SI7 in 40 (59.7%), SI13 in 44 (65.7%), and SI15 in 21 (31.3%). The patient with the highest mutation count showed predominant signatures related to MMR defect (SI6 and SI15), suggesting microsatellite instability-related hypermutation. However, the patient had no mutation in MMR-related genes detected by CANCERPLEX. Another patient who had the highest count in the UV-related SI7 (reconstructed mutation count 4.56, 12.3%) had an occupational exposure of UV, as he was employed as a neon sign engineer. Both smoking habits, plotted as PYI, and PD-L1 TPS showed no relationship with the reconstructed mutation counts.
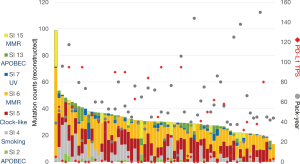
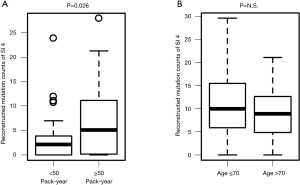
To determine whether the mutational signatures truly reflect the clinical characteristics, the relationships between PYI and reconstructed SI4 (smoking-related) mutation count and between age and reconstructed SI5 (clock-like) mutation count were analyzed. As expected, the SI4 mutation count was significantly higher in patients with high PYI (≥ median PYI, 50 PYI) than in patients with low PYI (<50 PYI, P=0.026, Figure 2A). In contrast, the SI5 mutation count was not related with the age of the patient (Figure 2B). No other significant correlations between clinical characteristics and signature were observed.
We next analyzed the signature that was most related to TMB. Reconstructed total mutation count and reconstructed signature count of each signature were plotted. The mutation counts of SI6 and SI15 were integrated, as well as those of SI2 and SI13, because of the hieratical similarities of each signature. SI4 was the most correlated with total mutation count, with a correlation coefficient of 0.51 (P<0.0001, Figure 3A). Both SI2/13 (APOBEC-related) and SI6/15 (MMR-related) had trends of positive correlations with total mutation count (P=0.07 and 0.01, respectively), but the correlation was weak (correlation coefficient =0.22 and 0.30, respectively; Figure 3B,C).
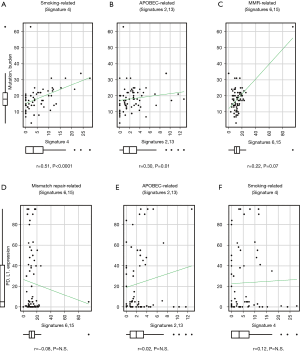
To identify the signature that was important for PD-L1 expression in the tumor, mutagenesis-related signatures were plotted along with PD-L1 TPS. No signature was related with PD-L1 expression (Figure 3D,E,F).
Discussion
Lung squamous cell carcinomas, especially those in patients with a smoking history, develop by the accumulation of multiple mutations including in TP53 (14,15). Many studies have shown that tobacco-related mutagens like benzo(a)pyrene cause G:C to T:A transversions by forming DNA adducts in the lung (16-18).
In this study, we referred to mutational signatures version 2, which revealed 30 distinct signatures based on an analysis of 10,952 exomes and 1,048 whole genomes across 40 distinct types of human cancer (19). The recently updated mutational signatures version 3, extracted from the 2,780 whole-genome variant calls produced by the ICGC/TCGA Pan Cancer Analysis of Whole Genomes Network, also described the signature derived from tobacco smoking as SBS4, which shows an identical pattern to SI4 in version 2 (20).
Deciphering mutational signatures from targeted sequencing is challenging, because the target region is narrow compared with whole exome sequencing (WES) or whole genome sequencing and the base triplet distribution of a target region could be different from what is available in whole genome sequencing. In fact, the percentages of SI4 in our samples were relatively low compared with the previous report (6), although estimated mutation counts belonging to SI4 did correlate with PYI. This implies that it is difficult to rank or compare mutational signatures that are reconstructed from targeted sequencing data within a single sample. As the techniques and cost of next-generation sequencing are rapidly improving, this issue will be resolved when whole exome (or whole genome) sequencing becomes more available for clinical use.
Most patients had more than four signatures within each tumor. Our results indicated that SI4 was most related with total mutation count, but other signatures also contributed to total mutation count. Tobacco mutagen is reported to induce mutations in TP53, which results in many other genomic alterations, causing cancer initiation and progression (15). Yoshino et al. reported no survival difference in patients with lung squamous cell carcinoma divided according to PYI (0, 1–20, 20–50 and 50+), whereas a significant difference in survival was observed in patients with lung adenocarcinoma divided according to PYI (21). When we compared estimated mutation counts belonging to SI4, patients with PYI more than 50 had more SI4 counts than patients with PYI 50 or below, suggesting that tobacco mutagen may continue to influence total mutation count even after a tumor has been established.
We detected some unique features by comparing signatures between samples. The patient with the highest mutation counts may likely have microsatellite instability, whereas no mutations were found in MMR enzymes. Another patient with a history of occupational exposure to UV had predominant SI7. Occupational exposure to UV is a risk for skin malignancy, but not for lung malignancy; there could have been an occult skin malignancy in that patient. Similarly, APOBEC mutagenesis has been related to human papillomavirus-associated carcinomas. However, it is difficult to distinguish whether the lung tumor is primary or metastatic, because previous studies did not distinguish between HPV-positive and -negative tumors (22).
We did not observe a relationship between PD-L1 expression and each mutational signature in this cohort. The importance of PD-L1 expression in regard to response to treatment with immune checkpoint inhibitors is controversial, especially in lung squamous cell carcinoma. Several reports have shown a correlation of TMB with clinical outcome (23). In our study, SI4 was the signature most related to total mutational burden. Several clinical studies have indicated that smoking history is related to good clinical outcome with immune checkpoint inhibitors (24). Further follow-up is needed to identify which signature is related to treatment outcome with immune checkpoint inhibitors.
Acknowledgments
We thank Gabrielle White Wolf, PhD, from Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing a draft of this manuscript.
Funding: This work was supported by funding from Denka Co., Ltd.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2602
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2602
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-2602
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2602). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Institutional Review Boards of both the Department of Surgery and Science, Graduate School of Medical Sciences, Kyushu University Hospital (IRB number: 674-01) and the Division of Thoracic and Cardiovascular Surgery, Niigata University Hospital (IRB number: 774). Written informed consent was obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ministry of Health, Labour and Welfare. Vital Statistics Japan. 2018. Available online: https://www.mhlw.go.jp/toukei/saikin/hw/jinkou/kakutei18/index.html
- Okamoto T, Takada K, Sato S, et al. Clinical and genetic implications of mutation burden in squamous cell carcinoma of the lung. Ann Surg Oncol 2018;25:1564-71. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246-59. [Crossref] [PubMed]
- UICC. The TNM classification of malignant tumors. 2009.
- Nagahashi M, Wakai T, Shimada Y, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med 2016;8:136. [Crossref] [PubMed]
- Blokzijl F, Janssen R, van Boxtel R, et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [Crossref] [PubMed]
- Takada K, Okamoto T, Toyokawa G, et al. The expression of PD-L1 protein as a prognostic factor in lung squamous cell carcinoma. Lung Cancer 2017;104:7-15. [Crossref] [PubMed]
- Ellrott K, Bailey MH, Saksena G, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 2018;6:271-81.e7. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 2000;77:81-137. [Crossref] [PubMed]
- Le Calvez F, Mukeria A, Hunt JD, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res 2005;65:5076-83. [Crossref] [PubMed]
- Ma B, Stepanov I, Hecht SS. Recent studies on DNA adducts resulting from human exposure to tobacco smoke. Toxics 2019;7:16. [Crossref] [PubMed]
- Pfeifer GP, Denissenko MF. Formation and repair of DNA lesions in the p53 gene: relation to cancer mutations? Environ Mol Mutagen 1998;31:197-205. [Crossref] [PubMed]
- Smith LE, Denissenko MF, Bennett WP, et al. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J Natl Cancer Inst 2000;92:803-11. [Crossref] [PubMed]
- Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402-7. [Crossref] [PubMed]
- Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94-101. [Crossref] [PubMed]
- Yoshino I, Kawano D, Oba T, et al. Smoking status as a prognostic factor in patients with stage I pulmonary adenocarcinoma. Ann Thorac Surg 2006;81:1189-93. [Crossref] [PubMed]
- Cannataro VL, Gaffney SG, Sasaki T, et al. APOBEC-induced mutations and their cancer effect size in head and neck squamous cell carcinoma. Oncogene 2019;38:3475-87. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Fujimoto D, Yoshioka H, Kataoka Y, et al. Efficacy and safety of nivolumab in previously treated patients with non-small cell lung cancer: a multicenter retrospective cohort study. Lung Cancer 2018;119:14-20. [Crossref] [PubMed]


