
RPB5-mediating protein promotes the progression of non-small cell lung cancer by regulating the proliferation and invasion
Introduction
The RNA polymerase II subunit 5 (RPB5)-mediating protein (RMP), also known as unconventional prefoldin RPB5 interactor (URI), is a member of the molecular chaperone prefoldin family. Many studies have confirmed that RMP plays an important role in cellular ubiquitination and transcription, which is mainly due to an interaction with subunit RPB5 of RNA polymerase II, but also partly due to interacting with other polypeptides to form protein complexes (1,2). The subunit RPB5 is one of the important targets in signal transducer and transcriptional regulation. It is closely related to specific binding targets of RNA polymerase II, such as Taf1 and RAP30, and thus participates in the transcriptional regulation and expression of downstream genes. Previous studies have shown that RMP is involved in the target of rapamycin (TOR) pathway, suggesting its possibility as a potential cancer-promoting gene (3-5). Our laboratory has revealed that RMP plays an important role in the hepatoma carcinoma cell proliferation, and overexpression of RMP can promote migration, invasion, and anti-apoptotic properties of HCC (6). Meanwhile, a relevant report of cholangiocarcinoma has indicated that RMP has a wide range of functions in the antioxidant process during the development of tumors (7).
Similarly, recent studies have identified RMP as an oncogene, and research has demonstrated that RMP is overexpressed in ovarian carcinoma, cervical cancer, and endometrial cancer. The role of RMP is catalytic during the occurrence and development of multiple myeloma (8-11). Furthermore, RMP participates in cytoplasmic signal transduction as well as intracellular signal transduction, which suggests the possibility of extensive involvement in carcinogenic processes (12).
Lung cancer is one of the leading causes of death worldwide. Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancer, mainly including squamous cell carcinoma (SCC), adenocarcinoma, and large cell carcinoma (13). The accumulation of genes and epigenetic changes is the underlying cause of lung cancer, leading to the activation of oncogenes and inactivation of tumor suppressor genes in tumor cells (14). Treatment of lung cancer includes surgery, radiotherapy, and chemotherapy, but the treatment effect is limited for the middle and late stages. The mutation of epidermal growth factor receptor (EGFR) and the dysregulated expression of various microRNAs in NSCLC have been reported in accumulating literatures. The high expression of RMP in liver cancer, ovarian cancer, cervical cancer, multiple myeloma, and endometrial carcinoma, has contributed to it being regarded as a universal biomarker. The expression of RMP in NSCLC has not yet been fully investigated. Our research group previously revealed that the expression of RMP in the NSCLC cell adenocarcinomic human alveolar basal epithelial cells (A549) is significantly higher than that in normal cells (15), suggesting that RMP may also play an important role in the development of NSCLC and has the potential to be developed into a therapeutic target.
In this study, the proliferation promotion of RMP on NSCLC cells were studied by cell counting kit-8 (CCK8). As in clinical practice, the NSCLC patients were widely treated with chemotherapy and radiotherapy, so we investigated whether RMP played a role in the radiosensitivity of NSCLC cells by CCK8 and flow cytometry. Furthermore, as the investigation of clinical samples revealed that RMP is correlated with tumor lymph nodes (LNs) and distant metastases, we studied the effect of RMP on the migration and invasion of NSCLC cell lines. Finally, we studied RMP in the tumor growth through the nude mouse transplantation model. At the same time, the expression of RMP, Bcl2-associated X protein (Bax), caspase-3, and B-cell lymphoma 2 (Bcl-2) in transplanted tumors were detected by immunohistochemistry (IHC), and it was further identified that RMP could play the role of tumor promotion by regulating the expression of apoptosis-related genes. This work may provide assistance in the creation of a theoretical basis for the diagnosis and therapy of NSCLC. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-3461).
Methods
Materials
A total of 20 pairs of human NSCLC tissues and corresponding adjacent tissues were provided by the Department of Thoracic Surgery, the First Affiliated Hospital of Soochow University, from March 2012 to June 2013 (all participants had not received treatment before the operation, and the project was approved by the participants and the ethics committee of First Affiliated Hospital of Soochow University). Informed consent was taken from all the patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
The cell lines A549 and PC-9 were provided by our laboratory, and the H226 and BEAS-2B cell lines were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. The plasmids RMP overexpression plasmid pCDNA-3.1, RMP interference plasmid pGPU6-RMPi, and RMP interference control plasmid pGPU6-SCR were constructed in advance by our laboratory. Roswell Park Memorial Institute (RMPI) 1640 medium, calf serum, fetal bovine serum (FBS), and Dulbeccos’ modified eagle medium (DMEM) medium were purchased from Invitrogen (Invitrogen Corp., Carlsbad, CA, USA). Trizol and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) kits were purchased from Stratagene (Stratagene Corp., San Diego, CA, USA). Western lysate, sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, bicinchoninic acid (BCA) protein quantitation kit, and SDS-PAGE preparation kit were purchased from Beyotime (Haimen, Jiangsu, China). The RMP antibody was provided by our laboratory, and other antibodies were purchased from Thermo (Thermo Fisher Scientific, Waltham, MA, USA). The CCK-8 kit was purchased from Dojindo (Dojindo Laboratories, Kumamoto, Japan). Bagg Albino (BALB)/C nude mice (5 weeks old) were purchased from the animal experimental center of Soochow University. Propidium iodide (PI) and Annexin V apoptosis kits were purchased from Thermo Fisher (Waltham, MA, USA). Polyvinylidene fluoride (PVDF) membrane was purchased from Bio-Rad company (Bio-Rad Laboratories, Hercules, CA, USA). The IHC staining kit was purchased from Abcam (Cambridge, UK). Gefitinib was purchased from the First Affiliated Hospital. The invasive test kit was purchased from BD company (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Cell culture
The NSCLC cells (A549, H226, and PC-9) and normal bronchial epithelial cells (BEAS-2B) were cultured in complete medium (DMEM) containing 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were maintained in a 37 °C incubator with 5% CO2. Medium changes, cell freezing, generation, cultivation of cells, and other operations were performed using a clean bench. Experiments were performed using cells in the logarithmic phase.
Human NSCLC tissue specimen preservation
All tumors and adjacent tissues were precisely diagnosed by an experienced pathologist and were frozen in liquid nitrogen.
Isolation of RNA
Logarithmic phase cells were harvested via trypsin. Tumor tissues were ground with a pestle and mortar in liquid nitrogen, and were then transferred to a 1.5 mL tube. Cells were washed with phosphate buffered saline (PBS), and then 1 mL of Trizol was added and mixed thoroughly. Next, 400 μL of chloroform was added and vortexed for 20 min. The mixture was incubated on ice for 5 min, and then centrifuged at 12,000 r/min, 4 °C, for 15 min. A pipette was used to add 450 μL supernatants to new, sterile, 1.5 mL RNase-free tubes, then 500 μL isopropanol was added to each tube, it was mixed by inversion, and 2–3 min later it was centrifuged at 12,000 r/min, 4 °C, for 15 min. The precipitation was collected and washed with 600 μL of 75% ethanol, and then centrifuged at 12,000 r/min, 4 °C, for 10 min. The precipitation was then collected and washed with 800 μL absolute ethanol, and centrifuged at 12,000 r/min, 4 °C, for 10 min. The resulting precipitation was air dried, redissolved in 20–50 μL diethy pyrocarboanate (DEPC).
Quantitative real-time PCR
The total RNA extraction and reverse transcription reactions were conducted using the PrimeScriptTM RT reagent Kit with gDNA Eraser Kit (Takara Bio. Inc., Kusatsu, Shiga, Japan). Real-time PCR analyses were conducted using ABI7500 Fast Real-Time PCR. The primers used for RMP were 5'-TGTCCCTCGCAAATCCATCCTG-3' (forward) and 5'-CTCCTCAAAACTCCCCGCCTA-3' (reverse); for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were 5'-GACCTGACCTGCCGTCTA-3' (forward) and 5'-AGGAGTGGGTGTCGCTGT-3' (reverse).
Western blot
Total proteins were extracted from cells using radio-immunoprecipitation assay (RIPA) lysis buffer containing 1% phenylmethylsulfonyl fluoride (PMSF). Proteins were separated on 10% SDS-PAGE and transferred to a PVDF membrane. Then, the membranes were blocked in 5% non-fat milk. After being washed in PBS with Tween 20 (PBST), membranes were incubated with 1:1,000 anti RMP primary antibody first at 4 °C overnight. After washing three times in PBST, the membranes were incubated with secondary antibody at room temperature for 1 h. The blot was detected by chemiluminescence.
Radiation procedure and drug treatment
Each group of cells was treated with 60Co gamma ray irradiation, with an absorbed dose efficiency of 1 Gy/min, which was performed at the radiology center of Soochow University. Cells were treated with 3, 6, 9, and 12 μg/mL of gefitinib, respectively.
Transient transfection
The A549 cells were divided into the following groups: control; pCDNA3.1/A549; pCDNA3.1-RMP/A549; pGPU6-scramble/A549; pGPU6-RMPi/A549. On the day before transfection, cells were inoculated in the 6 well plate at 4×105/well and cultured in the incubator at 5% CO2, 37 °C. When the confluence was 80–90%, cells were transfected with the indicated plasmids by liposomeTM 2000. The transfected cells were observed on fluorescence microscope, and the transfection efficiency was evaluated by qRT-PCR and western blot.
CCK8 assay
Cells were inoculated into 96 well plates at the density of 5×105/mL. Then, the CCK8 reagent was added to the well at 10 µL/well. Absorbance at 450 nm was measured after 3 h.
Apoptosis assayed by Annexin V/PI staining
Each group of cells (24 h after transient transfection) was irradiated with 12 Gy 60CO gamma rays. After 48 h, cells were digested and centrifuged at 2,000 r/min for 5 min. The cells were collected and washed by precooled PBS twice. Then, 100 µL of precooled PBS was added to resuspend the cells, and 10 µL Annexin V was added. Cells were cultured with Annexin V away from light on ice for 15 min. Then, 10 µL PI was added and cultured avoiding light at room temperature for 15 min. Fluorescence was detected by flow cytometry.
Cell cycle assay
Cell cycle was detected by PI single staining. Each group of cells (24 h after transient transfection) was irradiated with 12 Gy 60CO gamma rays. After 48 h, the cells were collected and fixed with 70% ethanol. Then, RNase A was added into the cell suspension and reacted for 30 min at room temperature. The addition of 5 µg PI into the cells was followed by incubation at room temperature for 30 min. Then, cells were detected by flow cytometry within 1 h.
Wound healing experiments
Cell migration was evaluated by wound healing experiments. After 24 h of transient transfection, cells were inoculated into 24 well plates. Another 24 h later, 100 µL pipette tips were used to create a width scratch. The scratch width was observed at 0, 24, and 48 h.
Invasion assay
Transwell was used to evaluate invasion ability. Non-invasive cells in the upper layer of the membrane were scraped, and the invading cells at the bottom of the membrane were immobilized (stained by 0.05% violet crystals). The invasive cells on the membrane were counted under optical microscope observation (×100).
Nude mice model
Experiments were approved by the authorization of the Animal Ethics Committee of Soochow University, and were performed in compliance with institutional guidelines for the care and use of animals. A total of 12 mice were divided into three groups: untreated A549 cells (n=4), pCDNA3.1-RMP/A549 cells (n=4), pGPU6-RMPi/A549 cells (n=4). Cells at a concentration of 5×106 in 0.1 mL PBS were injected subcutaneously into nude mice at the right anterior axilla. The tumor size and volume were measured once every two days (tumor volume = A×B2/2; A, the longest diameter; B, the shortest diameter). After 14 days, all mice were sacrificed uniformly, and the tumors were stripped for subsequent experiments.
Immunohistochemistry
Specimens from nude mice were fixed in 10% neutral-buffered formalin and embedded in paraffin. Sections were depleted of paraffin, rehydrated, and treated with H2O2 to block endogenous peroxidase activity. Each slide was incubated with primary antibodies for 1 h and subsequent secondary antibodies for 1 h at room temperature. Then, slides were reacted with horseradish peroxidase (HRP)-labeled streptavidin for 30 min at room temperature. Slides were stained using a 3,3’-Diaminobenzidine (DAB) kit.
Statistical analysis
All data are presented as mean ± SD. Statistics were analyzed using SPSS 18.0 software (IBM Corp., Armonk, NY, USA). Multivariate statistics were analyzed by analysis of variance (ANOVA). The mean between the two groups was analyzed by t-test. A P vale <0.05 was considered to be statistically significant.
Results
Expression of RMP in NSCLC cell lines
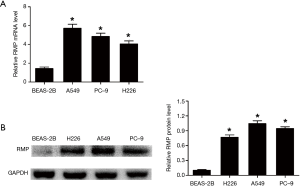
Normal bronchial epithelial cells BEAS-2B and three NSCLC cell lines A549, PC-9, and H226, were used to determine the mRNA level and protein level of RMP. The mRNA levels of RMP in the three NSCLC cells were significantly higher than in normal bronchial epithelial cells (*P<0.05) (Figure 1A). At the same time, Figure 1B shows that the protein levels of RMP in NSCLC cell lines were also higher than those in the BEAS-2B cell line (*P<0.05). These results indicated that RMP was highly expressed in the NSCLC cell lines, both at the transcriptional level and translational level, suggesting that it is a potential NSCLC- associated oncogene.
Expression of RMP in clinical NSCLC patients’ tissues and para-carcinoma tissues
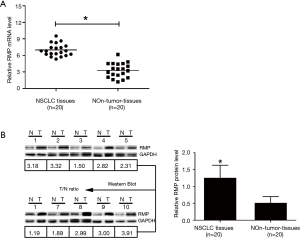
Given that the RMP level in NSCLC cells is increased, in order to further clarify the expression level of RMP in clinical NSCLC tissues, we detected RMP levels in 20 tissues collected from NSCLC patients. As shown in Figure 2A, the mRNA level of RMP in carcinoma tissues was higher than that in corresponding para-carcinoma tissues (*P<0.05). Similarly, as shown in Figure 2B, the protein level of RMP in carcinoma tissues was also higher than that in corresponding para-carcinoma tissues (ten representatives of the samples are shown). Statistics of the average protein level of RMP in 20 NSCLC patients are shown as a bar graph in Figure 2B. The results showed that protein levels of RMP in carcinoma tissues were significantly higher than those in para-carcinoma tissues (*P<0.05).
Correlation between RMP and clinicopathological characteristics of NSCLC
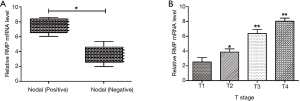
To further explore the correlation between RMP and clinicopathological features of NSCLC, we collected the patient clinical characteristics (Table 1). Correlations were detected between RMP and NSCLC LN status and T-stage status (*P<0.05) (Figure 3A,B). The results suggested that RMP might play a role in tumor recurrence and metastasis and RMP might act as a potential oncogene. Considering that the sample capacity was limited, future studies will need to be conducted on larger sample sizes to obtain more robust data.

Full table
The effect of RMP on the proliferation of A549 cells
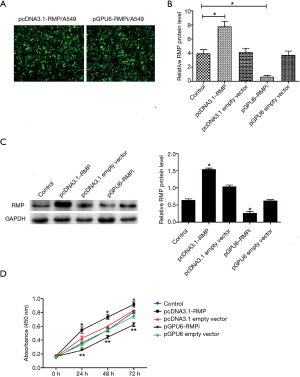
The A549 cell was transfected pCDNA3.1, pCDNA3.1-RMP, pGPU6, and pGPU6-RMPi by lipofectamineTM 2000. After 24 h of transfection, fluorescence was observed under the microscope (Figure 4A). The RMP mRNA of transfectants was detected by qRT-PCR (Figure 4B). Protein levels of RMP in transfectants were detected by western blot (Figure 4C). Statistics of the protein levels are shown in the bar graph. The results showed that pCDNA3.1-RMP/A549 expressed significantly higher RMP compared to the control, and pGPU6-RMPi significantly knocked down the expression of RMP.
Then, cell proliferation was determined by CCK8. As shown in Figure 4D, pCDNA3.1-RMP/A549 grew significantly faster than the control, and pGPU6-RMPi/A549 grew significantly slower than the control. The results suggested that RMP can promote cell proliferation.
The effect of RMP on the sensitivity to gefitinib and radiation of A549 cells
The A549 transfectants were treated with 3, 6, 9, and 12 μg/mL of gefitinib. Cell survival was determined by CCK8 (Figure 5A). As shown in Figure 5A, gefitinib treatment inhibited cell proliferation. The most resistance to gefitinib was shown by pcDNA3.1-RMP/A549, and pGPU6-RMPi/A549 was the most sensitive to gefitinib. The data suggested that RMP confers A549 resistance to gefitinib. Furthermore, after treatment with gefitinib, cell apoptosis was determined by flow cytometry (Figure 5B). As shown in Figure 5B, the percentage of cells under 10 µg/mL gefitinib treatment was 12.86% (control), 9.61% (pcDNA3.1-RMP), 12.82% (pcDNA3.1 empty vector), 23.68% (pGPU6-RMPi), and 12.15% (pGPU6 empty vector). Consistent with the results of CCK8, the apoptosis rate of pcDNA3.1-RMP/A549 was the lowest, and pGPU6-RMPi/A549 was the highest, when compared with the control group (*P<0.05), which also suggested that RMP confers A549 anti-apoptotic characteristics.
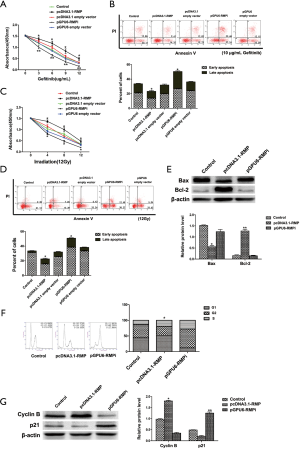
To further investigate the radiosensitivity of these A549 transfectants, cells were irradiated by 60Co gamma ray to undergo 4, 8, and 12 Gy radiation. Cell survival was assessed by CCK8 (Figure 5C). As shown in Figure 5C, under irradiation, pcDNA3.1-RMP/A549RMP survival rate was the highest and pGPU6-RMPi/A549 survival rate was the lowest, which suggested that overexpression of RMP conferred A549 cells resistant to irradiation. Cell apoptosis was assessed by flow cytometry (Figure 5D). Similarly, the apoptosis percentage of pcDNA3.1-RMP/A549 was the lowest, and pGPU6-RMPi/A549 was the highest. Then, apoptotic protein Bcl-2 and Bax levels were detected by western blot (Figure 5E). As shown in Figure 5E, the pro-apoptotic protein Bax level in pcDNA3.1-RMP/A549RMP significantly decreased, and anti-apoptotic protein Bcl-2 significantly increased in pGPU6-RMPi/A549. Some 48 h after irradiation, cell cycle after irradiation was further evaluated by flow cytometry (Figure 5F). After irradiation, the percentage of G2 stage in pcDNA3.1-RMP/A549 significantly decreased, which suggested that RMP alleviated G2 stage blockade induced by irradiation. Overexpression of RMP upregulated Cyclin B and downregulated p21 protein level, whereas RMP interference demonstrated the opposite effect (Figure 5G).
Effects of RMP on migration and invasion of A549 cells
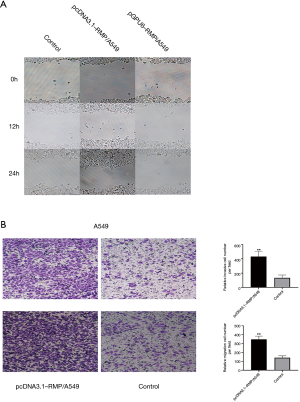
Migration and invasion are the main features of malignant lung cancer. A wound healing experiment was carried to evaluate the migration ability of A549 transfectants (Figure 6A). The transwell experiment evaluated the migration and invasion ability of A549 transfectants (Figure 6B). The results demonstrated that RMP conferred A549 cells stronger ability of metastasis and invasion.
Effect of RMP on the tumorigenicity of A549 cells
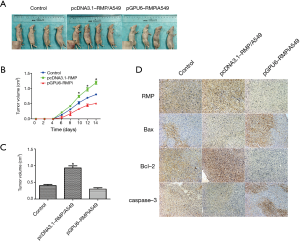
A nude mice model was used to investigate the tumor formation ability of A549 transfectants in vivo. A picture of the xenograft mice is shown in Figure 7A. The tumor growth is shown in Figure 7B; it shows that pcDNA3.1-RMP/A549 grew faster than the other two groups. The tumor weight is shown in Figure 7C. Tumors formed by pcDNA3.1-RMP/A549 were significantly larger than those of the other two groups (*P<0.05). The IHC staining of RMP, Bax, Bcl-2, and caspase-3 showed that in tumors formed by pcDNA3.1-RMP/A549, RMP and Bcl-2 expression increased, while Bax and caspase-3 expression decreased (Figure 7D). The changes of these molecules were opposite in the tumors formed by pGPU6-RMPi/A549 (Figure 7D).
Discussion
The carcinogenesis of NSCLC, the most common type of lung cancer, is a multifactorial, and involves a variety of gene expressions and physiological changes (16). A number of genes, such as PDCD4, PTEN, and miR-21, have been shown to be involved in the development of NSCLC when they are disordered (17-20). In this study, we focused on the interaction between RMP and NSCLC. It was shown that RMP could promote the proliferation of NSCLC cells in nude mice, and it was proven that the RMP promotes proliferation. Chemotherapy and radiotherapy are comprehensive methods for the treatment of NSCLC patients without EGFR or Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations. This work showed that RMP could decrease the sensitivity of NSCLC cells to chemotherapy or radiotherapy. At the same time, RMP can promote the expression of anti-apoptosis genes and inhibit the expression of apoptosis genes to reduce apoptosis induced by chemotherapy or radiotherapy. The X-ray irradiation can generally arrest cells in G2/M phase, thus slowing cell growth. This work showed that RMP could reverse cell cycle arrest by regulating Cyclin B and p21 expression, leading to the promotion of cell mitosis and accelerating cell growth. Moreover, RMP also demonstrated the ability to promote metastasis and invasion.
Although the effect of RMP on NSCLC biological characteristics has been widely studied, the exact mechanism by which it promotes NSCLC is still not fully understood. Recently, there was a report about the mTOR-S6K1 signal pathway in NSCLC. The mTOR expression level is significantly upregulated in lung cancer tissues, and rapamycin (the inhibitor of mTOR) effectively inhibits lung cancer cell proliferation (21). It has thus been identified that this is the exact mechanism of RMP in ovarian tumor progression (12,22). By combining the mechanism of RPM in ovarian cancer and the mTOR overexpression in lung cancer, it can be inferred that much interaction exists between RMP and the mTOR-S6K1 signaling pathway in NSCLC, which incited us to investigate the exact mechanism of RMP in NSCLC development. The study performed by the Shanghai Changzheng Hospital also showed that in multiple myeloma, RMP can promote STAT3 expression, which induces interleukin (IL)6 expression through interacting with nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) p65 (11). It is clear that the study of RMP in the development of NSCLC will contribute to the progress of NSCLC therapy. In this paper, we used qRT-PCR, western blot, and other classical methods to show that the expression of RMP in NSCLC cells and tissues was significantly higher than that in the control group. The expression of Bax and Bcl-2 can indicate apoptosis is well known. Higher levels of Bax promote apoptosis, but higher levels of Bcl-2 block apoptosis to support cell survival (6). In our manuscript, the IHC staining showed that the expression of RMP and Bcl-2 increased, and the expression of Bax and caspase-3 decreased in tumors. After irradiation, the pro-apoptotic protein Bax level in pcDNA3.1-RMP/A549RMP significantly decreased, and anti-apoptotic protein Bcl-2 significantly increased in pGPU6-RMPi/A549. Analysis by synthesis, RMP can promote the proliferation and migration of NSCLC by regulating the levels of Bax and Bcl-2, additionally, RMP can reduce the sensitivity of irradiation. The result of flow cytometry showed that RMP alleviated G2 stage blockade induced by irradiation. Meanwhile, when we analyzed the patients’ clinicopathological features, we found that RMP was correlated to LNs and T-stage status in NSCLC. All these results suggested that RMP might be a tumor-promoting gene associated with NSCLC.
However, the limitation of small sample size in this study means that we still need to collect more tissues to conduct further experiments to validate our findings. At present, the expression level of RMP in NSCLC has been determined in the prophase of this experiment, but its effect on biological characteristics of NSCLC and its mechanism of promoting cancer have not been clarified. Thus, the next step will be to study the biological characteristics of RMP in NSCLC from many aspects, and to demonstrate the carcinogenesis of RMP, specifically in an in vivo experiment. Through these specific studies, it may be indicated that RMP could be used as a target for the diagnosis and treatment of NSCLC, to improve the prognosis and survival rate of NSCLC patients.
Acknowledgments
Funding: This work was funded by the National Natural Science Foundation of China (81800279) and the Natural Science Foundation of Jiangsu Province (BK20180197).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-3461
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-3461
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-3461). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The project was approved by the participants and the ethics committee of First Affiliated Hospital of Soochow University (2018011). Informed consent was taken from all the patients. Animal experiments were approved by the Animal Ethics Committee of Soochow University, and were performed in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dorjsuren D, Lin Y, Wei W, et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol 1998;18:7546-55. [Crossref] [PubMed]
- Barnes DG, Vidiassov M, Ruthensteiner B, et al. Embedding and publishing interactive, 3-dimensional, scientific figures in Portable Document Format (PDF) files. PLoS One 2013;8:e69446. [Crossref] [PubMed]
- Wei W, Dorjsuren D, Lin Y, et al. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J Biol Chem 2001;276:12266-73. [Crossref] [PubMed]
- Washington K, Ammosova T, Beullens M, et al. Protein phosphatase-1 dephosphorylates the C-terminal domain of RNA polymerase-II. J Biol Chem 2002;277:40442-8. [Crossref] [PubMed]
- Delgermaa L, Hayashi N, Dorjsuren D, et al. Subcellular localization of RPB5-mediating protein and its putative functional partner. Mol Cell Biol 2004;24:8556-66. [Crossref] [PubMed]
- Yang H, Gu J, Zheng Q, et al. RPB5-mediating protein is required for the proliferation of hepatocellular carcinoma cells. J Biol Chem 2011;286:11865-74. [Crossref] [PubMed]
- Wan ZH, Jiang TY, Shi YY, et al. RPB5-Mediating Protein Promotes Cholangiocarcinoma Tumorigenesis and Drug Resistance by Competing With NRF2 for KEAP1 Binding. Hepatology 2020;71:2005-22. [Crossref] [PubMed]
- Theurillat JP, Metzler SC, Henzi N, et al. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell 2011;19:317-32. [Crossref] [PubMed]
- Gu J, Li X, Liang Y, et al. Upregulation of URI/RMP gene expression in cervical cancer by high-throughput tissue microarray analysis. Int J Clin Exp Pathol 2013;6:669-77. [PubMed]
- Gu J, Liang Y, Qiao L, et al. Expression analysis of URI/RMP gene in endometrioid adenocarcinoma by tissue microarray immunohistochemistry. Int J Clin Exp Pathol 2013;6:2396-403. [PubMed]
- Fan JL, Zhang J, Dong LW, et al. URI regulates tumorigenicity and chemotherapeutic resistance of multiple myeloma by modulating IL-6 transcription. Cell Death Dis 2014;5:e1126. [Crossref] [PubMed]
- Djouder N, Metzler SC, Schmidt A, et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell 2007;28:28-40. [Crossref] [PubMed]
- Sakamoto T, Furukawa T, Lami K, et al. A narrative review of digital pathology and artificial intelligence: focusing on lung cancer. Transl Lung Cancer Res 2020;9:2255-76. [Crossref] [PubMed]
- Lønvik K, Sørbye SW, Nilsen MN, et al. Prognostic value of the MicroRNA regulators Dicer and Drosha in non-small-cell lung cancer: co-expression of Drosha and miR-126 predicts poor survival. BMC Clin Pathol 2014;14:45. [Crossref] [PubMed]
- Yang S, Wang H, Guo Y, et al. RMP plays distinct roles in the proliferation of hepatocellular carcinoma cells and normal hepatic cells. Int J Biol Sci 2013;9:637-48. [Crossref] [PubMed]
- Schreurs O, Karatsaidis A, Balta MG, et al. Expression of keratins 8, 18, and 19 in epithelia of atrophic oral lichen planus. Eur J Oral Sci 2020;128:7-17. [Crossref] [PubMed]
- Ma XL, Liu L, Liu XX, et al. Prognostic role of microRNA-21 in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev 2012;13:2329-34. [Crossref] [PubMed]
- Zhu L, Liang J, Xia B, et al. Identification of somatic copy number variations in plasma cell free DNA correlating with intrinsic resistances to EGFR targeted therapy in T790M negative non-small cell lung cancer. J Thorac Dis 2020;12:883-92. [Crossref] [PubMed]
- Li Y, Zhang D, Chen C, et al. MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol Biol Cell 2012;23:1423-34. [Crossref] [PubMed]
- Liu XH, Lu KH, Wang KM, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer 2012;12:348. [Crossref] [PubMed]
- Pende M, Um SH, Mieulet V, et al. S6K1(-/-)/S6K2(-/-)mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol 2004;24:3112-24. [Crossref] [PubMed]
- Gstaiger M, Luke B, Hess D, et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 2003;302:1208-12. [Crossref] [PubMed]
(English Language Editor: J. Jones)


