
Enhanced expression of FGF19 predicts poor prognosis in patients with non-small cell lung cancer
Introduction
Among cancers, lung cancer ranks first in incidence and is the leading cause of cancer-related death worldwide. According to the GLOBOCAN estimate for 2018 (1), there were approximately 2.1 million newly diagnosed lung cancer cases and almost 1.67 million deaths, making lung cancer the leading cause of global cancer-related fatalities. In China, lung cancer has also gradually become a major public health concern (2), and although many treatment options and strategies exist, the situation is still not promising in the country.
Non-small cell lung cancer (NSCLC) accounts for the majority of lung cancer cases (3). Although radical surgery and chemotherapy remain the cornerstones for the treatment of NSCLC, a significant portion of patients still suffer postoperative recurrence. Major advances in the treatment of NSCLC, including molecular biology and immunotherapy, are gradually changing the received approach toward treatment (3). Despite these developments, many problems remain unresolved. First, identifying new biomarkers that can effectively predict postoperative recurrence risk remains an important problem for both physicians and patients. Secondly, which biomarkers can be used to reasonably guide prognosis and predict therapy response in NSCLC patients is a subject that is still being heavily researched. In other words, there is an urgent need to discover predictive biomarkers for identifying those NSCLC patients who can most benefit from complete resection and to identify biomarkers that can predict treatment efficacy.
The family of fibroblast growth factors (FGFs), which regulate a series of biological functions and multiple developmental processes, consists of 22 members divided into 7 subfamilies (4-6). The canonical FGFs account for the majority of FGF’s known functions in paracrine protein and autocrine protein. Another subgroup, endocrine FGFs (FGF19, FGF21, and FGF23), act as circulating hormones that differ from the canonical FGFs (6). As a member of the FGF family endocrine subgroup, FGF19 has a key role both in the regulation of bile acid homeostasis and the metabolism maintenance of protein and glucose (7). FGF19 is also critical for maintaining whole-body homeostasis and has therapeutic potential in malignant tumors (6). Acting as a circulating hormone (4), FGF19 has the potential to bind to four types of tyrosine kinase FGF receptors (FGFR 1–4) with quite a low affinity. Activation of canonical FGFs binding to FGFR4 requires heparin/heparin sulfate (HS) (4), while FGF19 requires β-klotho (KLB) as a coreceptor (7-9). Once FGF19 binds to FGFRs and coreceptor KLB, it triggers dimerization and autophosphorylation, ultimately activating the cytoplasmic signal transduction pathway (10).
In addition to its function in glucose metabolism and the regulation of bile acid homeostasis, FGF19 also participates in tumorigenic activity by activating FGF19-FGFR4 pathway, and is particularly involved in the pathogenesis of hepatocellular carcinoma cells (HCC) (11). The main downstream signaling pathways active in FGF19-FGFR4 include PI3K-AKT, Ras-Raf-MAPK, epithelial-mesenchymal transition (EMT), and the signal transducer and activator of transcription (STAT) pathway (5). Recent research has indicated that FGF19 enhances EMT in HCC by regulating FGFR4 activation through the GSK3β/β-catenin signaling cascade (12). Furthermore, certain drugs such as lenvatinib have demonstrated potential efficacy in anticancer therapy. Lenvatinib was shown to induce histological focal necrosis in Hep3B2.1-7 xenograft tumors harboring FGF19 overexpression and to promote the death of HCC cells by downstream regulation of FGF signaling pathway (13). Another study has identified FGF19 as a prognostic marker and potential driver gene of lung squamous cell carcinoma (LUSC) in Chinese patients who smoke (14).
Thus far, the correlation between FGF19 and postoperative recurrence risk or overall survival (OS) of NSCLC has not been fully studied. Therefore, we conducted this research to assess the value of FGF19 in NSCLC treatment and to ascertain the correlation between FGF19 expression and the clinicopathological characteristics of patients. Encouragingly, we found the upregulated expression of FGF19 to be associated with poor prognosis in NSCLC, which indicates that FGF19 may be a prognostic marker in NSCLC patients.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-126).
Methods
Bioinformatics analysis based on The Cancer Genome Atlas (TCGA) database
FGF19 messenger RNA (mRNA) expression data and corresponding survival data of patients with NSCLC were obtained from the official website of TCGA (https://portal.gdc.cancer.gov/). The lung adenocarcinoma (LUAD) dataset contains 469 tumor tissues and 58 normal tissues, while the lung squamous cell carcinoma (LUSC) dataset contains 490 cancer tissues and 51 normal tissues, with all the data being accompanied by clinical information. The “survival” and “survminer” packages of R software (R Foundation for Statistical Computing, Austria) were used to further process the RNA expression data and the survival data.
Patients
Specimens obtained from 187 NSCLC patients who underwent radical surgery for NSCLC from January 2010 to June 2016 were retrospectively analyzed in this study. The eligibility criteria were as follows: (I) histologically confirmed as primary NSCLC; (II) age ≥18 years; (III) well preserved tumor specimens accessible and available for immunohistochemistry (IHC) staining assay; (IV) clinical stage I–III according to the seventh edition of TNM classification; (V) integrated medical record and follow-up data of OS; (VI) no history of secondary primary tumors. This study was approved by the respective ethics committees of Nantong Tumor Hospital and the Affiliated Hospital of Nantong University. All patients or their family members provided written informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
IHC staining
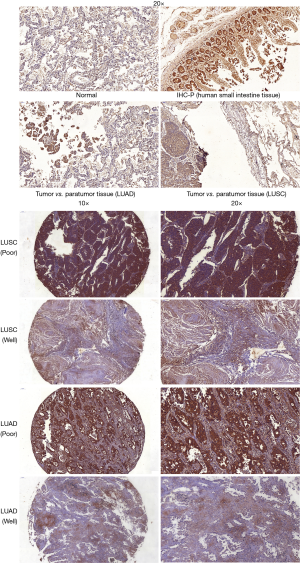
Tissue microarrays (TMAs) were constructed from formalin-fixed paraffin-embedded tumor tissues. The 4 µm tissue cores of approximately 70 donor blocks embedded in formalin-fixed paraffin were precisely arranged on a new paraffin block by a pathologist at the Affiliated Hospital of Nantong University. The expressions of FGF19 in the TMAs were detected by IHC assay. Anti-FGF19 antibody (1:100; Abcam, Cambridge, UK) and goat polyclonal to rabbit immunoglobin G (IgG) (1:10,000; Abcam) were used in the IHC assay. All procedures were performed according to the application manuals of the antibody. Before the experiment began, the antibody specificity of FGF19 was verified on formalin-fixed and paraffin-embedded human small intestine tissue according to the instructions of the antibody manufacturer.
After deparaffinization and rehydration, endogenous peroxidase activity was blocked in 3% hydrogen peroxide and left to react with methanol for 20 minutes. After a wash with phosphate-buffered saline, antigen retrieval was performed with a 1% concentration of Target Retrieval Solution Low pH (TRS Lo 50x, Dako Glostrup, Denmark) at 98 °C for 20 minutes. The slide was cooled at room temperature, and then the slides were incubated with the first anti-FGF19 antibody, ab225942 (dilution 1:100) at room temperature for 2 hours. Subsequently, the sections were incubated with a secondary antibody of goat polyclonal to rabbit IgG at a 1:10,000 dilution for 30 minutes at room temperature. Informed consent for use of human tissues was given by the patients and approved by the clinical research ethics committees of both hospitals.
IHC scoring
The semiquantitative scoring of IHC staining was independently scored by two pathologists. The expression of antibody in tumor cells was evaluated under a microscope at 200× magnification. Five high-power fields of view were randomly selected for the calculating of tissue cells. The density and intensity of positively stained cells were assessed by counting 100 tumor cells in each selected field. The intensity of staining was scored as follows: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. According to the density of staining and percentage of tumor cells, the scores were divided into four categories: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The combined score was then calculated by multiplying the intensity and density of staining (ranging from 0 to 12). For statistical analysis, a comprehensive score <6 was defined as a low expression, and a score ≥6 was defined as a high expression. Any disagreement between the two pathologists concerning the evaluation could be solved through consensus and reassessment.
Clinical and survival data extraction
Clinical data were extracted from the medical record, and included clinical and pathological information of age, sex, smoking and alcohol history, TNM stage, tumor size, pathological grade of tumor, vascular thrombus, neural invasion, and pleural invasion, and treatment history. Survival data were collected from follow-up records to evaluate the OS. The last follow-up date was December 31, 2019, and OS was defined as the time from initial therapy to death.
Statistical analysis
The clinical database was established using SPSS 25.0 software (IBM Corp., USA). The independent t-test and chi-square tests were performed for comparisons among groups. The cumulative survival rate was calculated by the Kaplan-Meier method, and between-group comparisons were calculated by the log-rank test. The Cox regression model was used to determine the risk factors of OS through univariate and multivariate analysis. All factors that showed statistical significance in univariate analysis (P<0.05) were included in multivariate analysis to identify independent risk factors. All statistical analyses were two-sided, and a P value <0.05 was considered statistically significant.
Results
NSCLC tissues exhibited elevated FGF19 mRNA expression
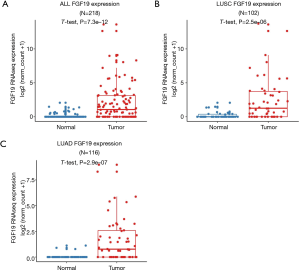
A detailed analysis was conducted using the data from TCGA database. The FGF19 mRNA expression levels in the NSCLC tissues and normal tissues were compared by t-test. The NSCLC tissues exhibited significantly higher levels of FGF19 mRNA expression than did the normal tissues (P<0.0001) (Figure 1A). Furthermore, we carried out a stratified analysis of the LUAD and LUSC subtypes. The FGF19 mRNA levels of both the LUAD tissues and LUSC tissues were notably higher than those of normal tissues (P<0.0001; Figure 1B,C).
Correlation of FGF19 mRNA levels with OS in NSCLC patients
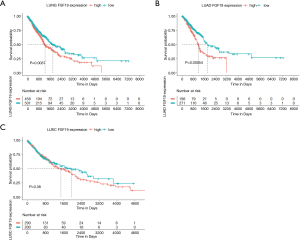
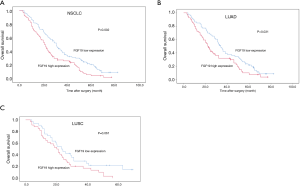
NSCLC patients with high FGF19 mRNA levels had lower OS than those with low expression (P<0.01; Figure 2A). Patients with high FGF19 expression in the LUAD cohort had shorter OS compared to those with low expression (P<0.001; Figure 2B); however, no significant difference was observed in the LUSC group (P=0.38; Figure 2C).
The expression of FGF19 correlated with copy number variation amplification
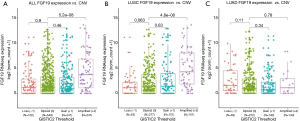
The standardized expression data of FGF19 and the segmentation data of copy number variation (CNV) were downloaded from TCGA database (https://portal.gdc.cancer.gov/). The classification value of CNV was calculated using GISTIC 2.0 (GenePattern) based on TCGA CNV segmentation, which is defined as follows: –2 = homozygous deletion, 1 = hemizygous deletion, 0 = neutral/no change, 1 = gain, 2 = high level amplification. As shown in Figure 3, the amplification of FGF19 CNV in the NCSLC group and the LUSC subgroup was significantly higher than that in other groups (P<0.0001) (Figure 3A,B), but no difference was observed in the LUAD subgroup (Figure 3C).
Correlation of FGF19 CNV amplification with OS in NSCLC patients
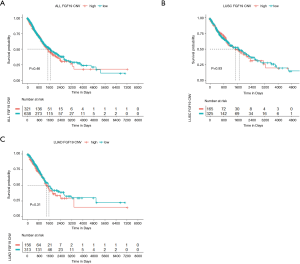
To determine the correlation of FGF19 CNV amplification with OS, we created two groups according to FGF19 CNV amplification: a high group (2) and a low group (−2,0,1). As shown in the Figure 4, FGF19 CNV amplification did not correlate with OS in NSCLC patients, either in the LUAD or LUSC subgroups.
FGF19 expression by IHC assay in NSCLC patients
FGF19 staining was mainly localized in the membrane and cytoplasm. In addition, positive staining was also detected in the nucleus of tumor cells. Examples of the FGF19 IHC staining of different tissue types are shown in Figure 5.
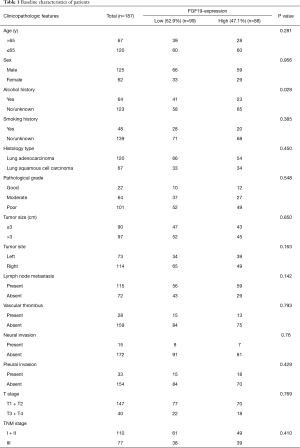
Characteristics of NSCLC patients
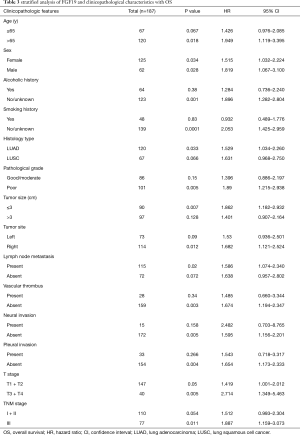
Patient characteristics and pathological features are shown in Table 1. All 187 NSCLC patients underwent radical surgery. Based on the combined FGF19 IHC score, 88 of 187 patients showed high FGF19 expression and 99 patients showed low FGF19 expression. The distribution of various clinical features was relatively balanced, and, as expected, there was no obvious difference.
Full table
Increased expression of FGF19 by IHC assay predicted poor OS in NSCLC patients
Additionally, the overexpression of FGF19 predicted poor OS in NSCLC patients (P=0.002) (Figure 6A). Separate analysis of LUAD and LUSC patients showed different outcomes: LUAD patients with high FGF19 expression had shorter OS than those with low expression (P=0.031, Figure 6B), while no difference was observed in LUSC patients (P=0.061, Figure 6C).
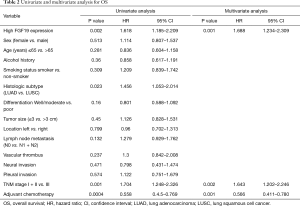
Survival outcomes and COX regression analysis of prognostic factors
In order to explore the prognostic significance in NSCLC, we analyzed the relevant pathological factors and clinical features (Table 2). Among the clinicopathological factors, high FGF19 expression, LUAD, TNM stage, and adjuvant chemotherapy were significantly associated with worse OS in the univariate analysis (P<0.05; Table 2). Furthermore, multivariate analysis was performed to determine the prognostic value of FGF19 expression using the Cox proportional hazards model. The significant factors from univariate analyses, including high FGF19 expression, TNM stage, and adjuvant chemotherapy, were selected for multivariate analysis. COX regression analysis revealed that high FGF19 expression [hazard ratio (HR) =1.688, 95% confidence interval (CI): 1.234–2.309, P=0.001), TNM stage (HR =1.643, 95% CI: 1.202–2.246; P=0.002), and adjuvant chemotherapy (HR =0.566, 95% CI: 0.411–0.780; P=0.001) were independently associated with OS. These results indicated that FGF19 expression was an independent prognostic factor and that overexpression of FGF19 was associated with poor OS.
Full table
Correlation of FGF19 expression and clinicopathological characteristics associated with OS (stratified analysis)
The correlation between FGF19 expression and the clinicopathological characteristics related to OS was analyzed by Cox regression analyses. As listed in Table 3, the elevated FGF19 expression and the clinicopathological characteristics related to OS were significantly associated with older age (>65 years), absence of alcohol history, absence of smoking history, LUAD, poor pathological grade, tumor size (≤3 cm), right-side tumor site, lymph node metastasis, absence of vascular thrombus, absence of neural invasion, absence of pleural invasion, T3–T4 stage, and TNM stage III.
Full table
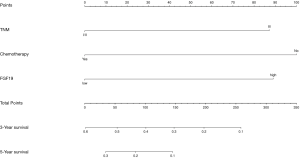
Prognostic nomograms for OS of NSCLC
Significant independent factors based on the multivariate analysis, including FGF19 expression, TNM stage, and adjuvant chemotherapy, were included in the nomogram to predict the 3- and 5-year survival of the NSCLC patients after radical surgery. The concordance index of the nomogram was 0.636 (Figure 7).
Discussion
Genetic changes of the FGF19 gene have been found in many types of malignant tumors, including HCC (15) and head and neck squamous cell carcinoma (HNSCC) (16). However, the role of FGF19 in the prognosis of NSCLC after surgery has thus far not been examined. Although postoperative adjuvant treatment of NSCLC has become standard care over the past few decades, the surgical outcome for early-stage NSCLC is still unsatisfactory (17). Therefore, identifying whether or not FGF19 is an oncogene and pivotal driver of NSCLC can provide potential opportunities for targeted intervention.
The present study found that (I) the expression of FGF19 in NSCLC tissues was higher than that in normal tissues, regardless of the level of mRNA or protein. (II) Cox proportional hazards regression analyses revealed that high FGF19 expression, TNM stage, and adjuvant chemotherapy were independently associated with OS. Our results thus provide clinical evidence that FGF19 plays an essential role in the prognosis of NSCLC.
The crucial roles of the endocrine FGF19 subfamily in glucose, bile acid, and phosphate homeostasis have been extensively studied in various disease, including malignant tumor. It is acknowledged that FGF19 regulates the metabolism of bile acids through the KLB receptor complex, and FGF19 has been reported to be associated with the occurrence of HCC (18). Other research has revealed (19) that FGF19 stimulates tumor progression by activating the STAT3 pathway and targeting FGF19-FGFR4 pathway, and this may provide a potential basis for improving the treatment of liver cancer. Another study has found that breast cancer cells with high expression of FGF19 can secrete higher levels of FGF19, thereby triggering multiple intracellular signaling pathways through the receptor (20). Moreover, FGF19 might be a key contributor to the autocrine FGF19/FGFR4 signaling pathway in HNSCC, and FGF19-based therapies have a promising future (16).
The discovery that FGFs regulate multiple of biological functions has precipitated the recent emergence of FGF19-based therapies as promising therapeutic approaches for the treatment of a variety of chronic diseases and malignant cancer (6,10,21). As an endocrine ligand, FGF19 has a more specific selective affinity to FGFR4 than other FGFR members, and thus various drugs targeting the FGF19-FGFR4 pathway (10,11,22) are being investigated. The current therapeutic options include anti-FGF19 mimics or variants, such as M70, specifically designed to eliminate FGFR4 binding (19). M70 is an engineered, non-tumorigenic FGF19 that preserves biological function but does not promote tumor formation. Research has attested to the potential of using it as a selective modulator to suppress FGF19-dependent tumor growth. With demonstrated efficacy in animal models and tumor patients, therapeutic approaches using FGFR4-specific inhibitors such as BLU9931 (18,23) and H3B-6527 (24), are paving the way in suppressing hyperactive FGF19-FGFR4 signaling in malignant tumors. In addition, non-specific FGFR inhibitors such as lenvatinib (E7080) (13,25) and ponatinib (26,27) might have a promising future in cancer treatment, but require more clinical and pharmacological testing. FGFR monoclonal antibodies directed against FGF or FGFR are also in clinical development (28). Several FGFR inhibitors that have entered early-phase trials shown the non-selectivity and side effects from pan-FGFR inhibition, mainly from the on-target toxicity and hyperphosphatemia (18). Further studies that will assess the safety issues and therapeutic effects of FGF19 are eagerly anticipated. Although many challenges will inevitably arise, the future of FGF19-based therapy is promising.
In our study we found the expression of FGF19 is significantly upregulated in NSCLC. Besides, we discovered that the expression of FGF19 correlated with CNV amplification and enhanced FGF19 expression predicted poor OS in NSCLC patients. In addition, stratified analysis found that the expression of FGF19 and the clinicopathological characteristics related to OS were significantly correlated with age, alcohol history, smoking history, LUAD, poor pathological grade, tumor size, lymph node metastasis, vascular thrombus, neural invasion, pleural invasion, T stage, and TNM stage, which indicates the joint prediction significance for patients with these clinicopathological characteristics. ThisOur research supports the value of FGF19 as an independent prognostic indicator for thosethese NSCLC patients undergoing surgical resection. Whereas, the problem of postoperative recurrence of NSCLC patients is also one of the key concerns of clinicians. We only explored the correlation of FGF19 with the overall survival due to the incomplete data of PFS, and this is the limitation of the study. As a member of an atypical subfamily of FGFs, the concentration of FGF19 can be detected in peripheral blood in both healthy people and the patients (6). Further exploration should focus on whether FGF19 could be used as a biomarker for early screening of NSCLC, the detective value of FGF19 in peripheral blood of NSCLC patients, and the correlation between FGF19 and recurrence of NSCLC. One recent study (29) explored the clinical outcomes and genomic data of 179 advanced epidermal growth factor receptor (EGFR)-mutated NSCLC patients undergoing first-line treatment of EGFR tyrosine kinase inhibitors (TKIs), and revealed that the amplification of FGF19 was significantly associated with poor clinical outcomes in these EGFR-mutated NSCLC patients. From the perspective of clinical doctors, future work should focus on exploring the resistance mechanisms and creating methods to reverse drug resistance in these EGFR-mutated patients with high FGF19 expression/amplification.
Conclusions
Our research demonstrated that FGF19 expression is significantly enhanced in NSCLC patients and that the overexpression of FGF19 is correlated with shorter OS in NSCLC, especially the LUAD subtype. FGF19 might therefore serve as a potential biomarker for predicting poor survival in NSCLC patients.
Acknowledgments
We especially thank Dr. Yanan Chen (3D Medicines Inc., Shanghai) and Dr. Shiqing Chen (3D Medicines Inc., Shanghai) for their assistance with bioinformatics analysis of TCGA.
Funding: This work was supported by the Scientific Research Subject of the Health Commission of Nantong (WKZL2018048), the Scientific Research Subject of Nantong Science and Technology Bureau (MSZ19233), Scientific Research Project of “333 Project” in Jiangsu Province (BRA2019030), and Nantong Science and Technology Foundation grant (MS22019008).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-126
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-126
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-126). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the respective ethics committees of Nantong Tumor Hospital and the Affiliated Hospital of Nantong University (No. 2016-094). All patients or their family members provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. [Crossref] [PubMed]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 2009;8:235-53. [Crossref] [PubMed]
- Liu Y, Cao M, Cai Y, et al. Dissecting the Role of the FGF19-FGFR4 Signaling Pathway in Cancer Development and Progression. Front Cell Dev Biol 2020;8:95. [Crossref] [PubMed]
- Degirolamo C, Sabba C, Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat Rev Drug Discov 2016;15:51-69. [Crossref] [PubMed]
- Lan T, Morgan DA, Rahmouni K, et al. FGF19, FGF21, and an FGFR1/beta-Klotho-Activating Antibody Act on the Nervous System to Regulate Body Weight and Glycemia. Cell Metab 2017;26:709-18.e3. [Crossref] [PubMed]
- Lang L, Teng Y. Fibroblast Growth Factor Receptor 4 Targeting in Cancer: New Insights into Mechanisms and Therapeutic Strategies. Cells 2019;8:31. [Crossref] [PubMed]
- Lee S, Choi J, Mohanty J, et al. Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling. Nature 2018;553:501-5. [Crossref] [PubMed]
- Hallinan N, Finn S, Cuffe S, et al. Targeting the fibroblast growth factor receptor family in cancer. Cancer Treat Rev 2016;46:51-62. [Crossref] [PubMed]
- Raja A, Park I, Haq F, et al. FGF19-FGFR4 Signaling in Hepatocellular Carcinoma. Cells 2019;8:536. [Crossref] [PubMed]
- Zhao H. FGF19 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells by modulating the GSK3β/βcatenin signaling cascade via FGFR4 activation Oncotarget 2016;7:13575-86. [Crossref] [PubMed]
- Hoshi T, Watanabe Miyano S, Watanabe H, et al. Lenvatinib induces death of human hepatocellular carcinoma cells harboring an activated FGF signaling pathway through inhibition of FGFR-MAPK cascades. Biochem Biophys Res Commun 2019;513:1-7. [Crossref] [PubMed]
- Tan Q. Identification of FGF19 as a prognostic marker and potential driver gene of lung squamous cell carcinomas in Chinese smoking patients. Oncotarget 2016;7:18394-402. [Crossref] [PubMed]
- Teng Y, Zhao H, Gao L, et al. FGF19 Protects Hepatocellular Carcinoma Cells against Endoplasmic Reticulum Stress via Activation of FGFR4-GSK3beta-Nrf2 Signaling. Cancer Res 2017;77:6215-25. [Crossref] [PubMed]
- Gao L, Lang L, Zhao X, et al. FGF19 amplification reveals an oncogenic dependency upon autocrine FGF19/FGFR4 signaling in head and neck squamous cell carcinoma. Oncogene 2019;38:2394-404. [Crossref] [PubMed]
- Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol 2010;28:35-42. [Crossref] [PubMed]
- Subbiah V, Pal SK. Precision Oncology for Hepatocellular Cancer: Slivering the Liver by FGF19-FGF4-KLB Pathway Inhibition. Cancer Discov 2019;9:1646-9. [Crossref] [PubMed]
- Zhou M, Wang X, Phung V, et al. Separating Tumorigenicity from Bile Acid Regulatory Activity for Endocrine Hormone FGF19. Cancer Res 2014;74:3306-16. [Crossref] [PubMed]
- Zhao X, Xu F, Dominguez NP, et al. FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol Carcinog 2018;57:1616-25. [Crossref] [PubMed]
- Repana D, Ross P. Targeting FGF19/FGFR4 Pathway: A Novel Therapeutic Strategy for Hepatocellular Carcinoma. Diseases 2015;3:294-305. [Crossref] [PubMed]
- Hui Q, Jin Z, Li X, et al. FGF Family: From Drug Development to Clinical Application. Int J Mol Sci 2018;19:1875. [Crossref] [PubMed]
- Hagel M, Miduturu C, Sheets M, et al. First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway. Cancer Discov 2015;5:424-37. [Crossref] [PubMed]
- Joshi JJ, Coffey H, Corcoran E, et al. H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma. Cancer Res 2017;77:6999-7013. [Crossref] [PubMed]
- Matsuki M, Hoshi T, Yamamoto Y, et al. Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models. Cancer Med 2018;7:2641-53. [Crossref] [PubMed]
- Katoh M. FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis Int J Mol Med 2016;38:3-15. (Review). [Crossref] [PubMed]
- Salgia R. Fibroblast growth factor signaling and inhibition in non-small cell lung cancer and their role in squamous cell tumors. Cancer Med 2014;3:681-92. [Crossref] [PubMed]
- Rijavec E, Genova C, Barletta G, et al. Investigational drugs targeting fibroblast growth factor receptor in the treatment of non-small cell lung cancer. Expert Opin Investig Drugs 2017;26:551-61. [Crossref] [PubMed]
- Cheng Y, Ma L, Liu Y, et al. Comprehensive characterization and clinical impact of concomitant genomic alterations in EGFR-mutant NSCLCs treated with EGFR kinase inhibitors. Lung Cancer 2020;145:63-70. [Crossref] [PubMed]
(English Language Editor: J. Gray)


