
Molecular characterization of advanced non-small cell lung cancer patients by cfDNA analysis: experience from routine laboratory practice
Introduction
Lung cancer, which is one of the most frequent and lethal cancers worldwide (1), has traditionally been classified, based on clinical-histopathological characteristics, into the non-small cell lung carcinoma (NSCLC) subgroup, including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma; and the small cell lung cancer (SCLC) subgroup. The NSCLC and SCLC subgroups correspond to 85% and 15% of all lung cancers respectively (2).
Recently, advances in the understanding of the molecular bases of lung cancer have promoted a new classification based on molecular profiling (2). Mutations in the epidermal growth factor receptor (EGFR) gene are common driver events in lung adenocarcinomas (3) with the prevalence of EGFR mutations ranging from 14.1% in European patients to 38.4% in Asian patients. Treatment with EGFR tyrosine kinase inhibitors (TKIs) is the standard-of-care in first-line therapy for NSCLC patients with EGFR mutant tumors. While several generations of TKIs have considerably improved the prognosis of the disease (4), patients ultimately develop resistance to TKI treatment. The acquisition of the p.T790M EGFR mutation is a common mechanism of resistance observed in 50% of patients treated with first and second-generation TKIs (5). Osimertinib, a third-generation TKI, is the standard of care in patients with an acquired p.T790M EGFR mutation. To date, several clinical guidelines recommend testing for EGFR mutations in all advanced NSCLC patients, independently of the clinical characteristics, to identify those patients most likely to benefit from targeted EGFR inhibition (6). In addition, a new biopsy is strongly advised at the time of progression to first-line EGFR TKIs to rule out the targetable p.T790M mutation (7).
Tumor tissue is the gold standard for the molecular characterization of NSCLC tumors. However, up to 60% of patients do not have a biopsy sample suitable for molecular analysis (8). Furthermore, tissue biopsies are obtained by invasive procedures that increase the risk of adverse events, and genotyping possibilities are limited due to inter- and intra-tumor heterogeneity (9). To overcome these limitations, circulating free DNA (cfDNA) in serum or plasma has emerged as an alternative and easily accessible sample source. Clinical guidelines recommend EGFR molecular testing of cfDNA from plasma in mutant EGFR lung cancer patients when resistance develops or at diagnosis when tissue availability is limited or difficult to obtain (7). EGFR mutation analysis in cfDNA can be carried out by distinct polymerase chain reaction (PCR) or next-generation sequencing (NGS) methods, and its clinical utility has mainly been evaluated in patients in clinical trials (10,11) or by analyzing synthetic DNA (12-14). Although different methods have shown differing sensitivity/specificity and diagnostic accuracy, EGFR testing in plasma has proven to be a reliable substitute for tissue testing (10,15-20). The cobas® EGFR Mutation Test v2 (cobas® EGFR Test) (Hoffman-La Roche Ltd, Basel, Switzerland) is an FDA and CE approved real-time PCR test currently used in the clinical setting to analyze specific target EGFR mutations in metastatic NSCLC patients (12,21). The test provides a semiquantitative index (SQI), which reflects a trend for the proportion of mutated versus wild-type copies of the EGFR gene in the cfDNA (8).
cfDNA is comprised of short nuclear DNA fragments of variable length that are released into plasma and other biological fluids by stromal and tumor cells (circulating tumor DNA; ctDNA). Although the biology of cfDNA is poorly understood, apoptosis and necrosis have been established as the major mechanisms of release. Higher concentrations of cfDNA have been found in lung cancer patients compared to healthy individuals (22,23). In addition, levels of both cfDNA and ctDNA have been associated with disease prognosis and relapse, highlighting their potential clinical utility (24,25).
The length of cfDNA fragments has been associated with the mechanism of their release. While apoptosis promotes the release of DNA fragments of approximately 180 base pairs (bp) or multiples thereof, necrosis produces larger fragments. Moreover, fragments between 1,000–3,000 bp are associated with active release processes (26). In contrast to total cfDNA levels, the significance of cfDNA fragmentation pattern has been less explored in the clinical context. Recent studies in advanced pancreatic cancer patients (27) and breast or prostate cancer patients (28), however, suggest that the cfDNA fragmentation pattern may provide diagnostic and prognostic value in cancer patients.
In this report, we evaluate the utility of molecular cfDNA characterization of advanced or metastatic NSCLC TKI-naïve patients in a routine clinical setting, using EGFR mutation testing and analysis of the concentration and fragmentation pattern of cfDNA.
We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-3142).
Methods
Patient enrollment
The study includes a prospectively recruited cohort of 173 NSCLC patients referred for plasma EGFR molecular testing at Hospital Clínic, Barcelona, between 2017 and 2020. At the time of inclusion, all patients were diagnosed with locally advanced or metastatic NSCLC and were TKI-naïve. Clinical data such as gender, age, tumor histology, disease stage, smoking status and molecular status of EGFR in tissue biopsy were obtained from their medical records.
The protocol was approved by the Hospital Clinic Ethics Committee (approval registration HCB/2016/0889) and was conducted in accordance with Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients.
Isolation of cfDNA from plasma
For cfDNA analysis, peripheral whole blood was collected from each subject in a 5 ml EDTA-K2 tube. When possible, two blood extractions were taken from each patient in order to evaluate the reproducibility of the test. After 15 to 20 minutes at rest in an upright position at room temperature, samples were centrifuged at 1,600 g for 10 minutes to collect 2 mL of plasma, which was transferred to a clean sterile tube. After a second centrifugation at 16,000 g for 10 minutes, plasmas were stored at −20 °C. The entire procedure is completed within three hours of blood extraction. The cfDNA was isolated using the cobas® cfDNA Sample Preparation Kit as per the manufacturer’s instructions.
Determination of cfDNA concentration and fragment size
The concentration of eluted cfDNA was measured using a Qubit 4 Fluorometer. The cfDNA fragment size for each sample was determined by microchip electrophoresis using an Agilent 2100 Bioanalyzer and the Agilent High Sensitivity DNA. The Agilent 2100 Bioanalyzer software was used to obtain the fraction of cfDNA and average size corresponding to apoptotic DNA fragments (100–250 and 250–700 bp). The fraction of cfDNA fragment size and cfDNA levels were obtained by individuals blinded to the clinical data to reduce potential bias.
EGFR mutation analysis
Analysis of EGFR mutations in cfDNA was performed by the cobas® EGFR Test. The test allows the detection of 42 recurrent point mutations, deletions, and insertions located in exons 18 to 21. The cobas® EGFR Test is a RT-PCR assay designed to detect G719X substitution mutations in exon 18, deletion mutations in exon 19, T790M and S768I substitution mutations in exon 20, insertion mutations in exon 20, and L858R and L861Q substitution mutations in exon 21. However, the specific mutation found in each sample is not displayed in the final test report. The cobas® z480 software reports a SQI value. Mutation status from tumor biopsy and the method used for the molecular characterization was obtained from medical records. Both plasma and tissue analysis were performed by individuals blinded to the clinical data.
Statistical analysis
Analysis of cfDNA concentration between groups was carried out by non-parametric rank tests; Kruskal-Wallis and Mann-Whitney U. Comparisons of the SQI of the two most prevalent mutations were made using the Student’s t-test. Correlation between SQI and cfDNA concentration was carried out using the Spearman rank correlation test. The chi square test was used to compare the prevalence of EGFR mutations between groups. A simple linear regression was made to determine the correlation coefficient between the two vials of the same patient.
To evaluate concordance in molecular status between tissue and plasma, only patients with both tissue and plasma samples analyzed with less than two months difference were selected. Sensitivity, specificity, positive predictive value, negative predictive value, global accuracy, and 95% confidence intervals were calculated as standard measures of diagnostic test validity. Results were considered statistically significant when the P value was <0.05. We calculate the predictive capability for distinguishing stage III and stage IV for the cfDNA concentration and integrity using receiver operating characteristic (ROC) curve analysis, by identifying the cut-off for both parameters for the detection of patients with stage IV lung cancer with 95% specificity and maximal sensitivity. All statistical analysis was performed using IBM SPSS software (version 23).
Results
Association of clinicopathological characteristics to cfDNA concentration and fragmentation pattern
We prospectively analyzed cfDNA from 173 advanced or metastatic NSCLC TKI-naïve patients. Among them, 80.9% (140/173) were diagnosed with lung adenocarcinoma (ADK) and 19.1% (33/173) with other histologies such as adenoid cystic carcinoma, pulmonary pleomorphic carcinoma, cancer of unknown primary, squamous cell carcinoma or NSCLC-non-otherwise specified (NSCLC-NOS). Stage III and IV represented 9.8% (17/173) and 90.2% (156/173) of cancer patients respectively. The demographic and clinical characteristics of the patients are shown in Table 1.
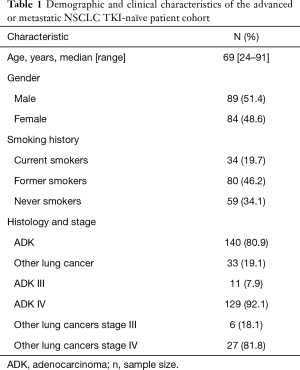
Full table
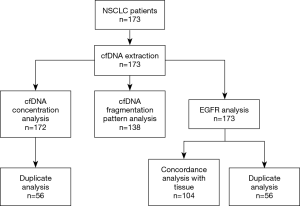
We assessed whether cfDNA concentration and fragmentation pattern were associated with the clinical characteristics of the patients (Table 2). The cfDNA concentration and fragmentation pattern were determined in 99.4% (172/173) and 79.8% (138/173) of patients respectively (Figure 1).
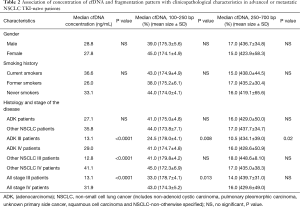
Full table
The median cfDNA concentration detected among patients was 28.3 ng/mL (range, 6.1–529). We did not find any difference in cfDNA concentration among EGFR mutant patients based on the type of EGFR mutation. We found significant differences in the cfDNA concentration among patients based on disease stage. Stage IV cancer patients showed higher concentrations of cfDNA compared to stage III patients (31 vs. 13.9 ng/mL; P<0.0001). Such differences were observed among patients with adenocarcinoma tumors as well as patients with other histological subtypes.
The analysis of the fragmentation pattern was split between the fraction of fragment sizes between 100–250 bp, corresponding to nucleosome fragments; and the fraction of fragment sizes between 250–700 bp, corresponding to polynucleosome fragments. All patients showed cfDNA in both size ranges with a median size of 175 and 438 bp in each range respectively. Overall, the stage IV cancer patients presented a higher fraction of 100–250 bp fragment size compared to stage III cancer patients (43.0% vs. 33.0%; P=0.01, Figure 2A). Among adenocarcinoma patients, stage IV patients showed a higher fraction of 100–250 bp fragment size but also 250–700 bp fragment size (Figure 2B). These differences were not observed among patients with other NSCLC tumors probably due to the reduced sample size.
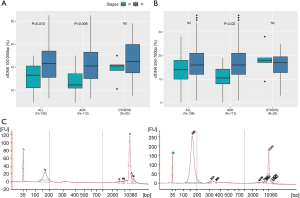
We evaluated the diagnostic utility of both cfDNA concentration and fraction of 100–250 bp fragment size as biomarkers for the disease stage in NSCLC patients. We performed ROC curve analysis to identify the cut-off value for both parameters with 95% specificity and maximal sensitivity. In our cohort of patients, 20.3 ng/mL of cfDNA concentration and 50.5% of 100–250 bp fragment size were the optimal cut-offs to detect stage IV cancer patients with a sensitivity of 72.3% for cfDNA concentration and 36.8% for fragmentation pattern. Although both parameters reached statistical significance, the cfDNA concentration showed higher AUC compared to the 100–250 bp fraction (0.86% vs. 0.71%) (Table 3).
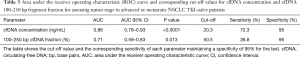
Full table
Characterization of EGFR mutations in cfDNA from plasma
We analyzed the presence of EGFR mutations in cfDNA in all patients. The median turnaround time for delivering the EGFR mutation status to the clinicians was 7 days. We detected EGFR mutations in 24.3% (42/173) of cases, corresponding to deletions in exon 19 (14.5% of cases), p.L858R mutation (7.5% of cases), mutations at codon p.G719X (1.2% of cases), mutations at codon p.L861Q (0.6% of cases) and Ex20Ins (0.6% of cases). All mutant EGFR tumors belonged to patients with adenocarcinoma, except for two patients in which the histological information of the tumor was not available.
We detected a higher prevalence of EGFR mutations in plasma from women vs. men (34.5% vs. 14.6%, P=0.002) and non-smokers vs. past or current smokers (49.2% vs. 11.4%, P<0.0001). In contrast, we found similar frequencies between different disease stages. EGFR mutations were found in 17.6% of stage III cancer patients and 25.0% of stage IV cancer patients.
We evaluated the reproducibility of the cobas® EGFR Test in 56 patients by analyzing two distinct vials obtained during the same blood draw. The cfDNA concentration showed a correlation coefficient of r2=0.99 between both vials (Figure 3A). Among EGFR mutant patients, the SQI value for the EGFR mutation showed a correlation coefficient of r2=0.70 between the vials (Figure 3B). We obtained the same molecular status of EGFR in both vials in 96.5% (54/56) of patients. The two remaining cases with discrepant results between the vials harbored the p.L858R EGFR mutation. The mutation was only detected in one of the vials, and the test reported a SQI value of 5.4 in one patient and an SQI value of 6.1 in the other patient. In both patients, the cfDNA concentration was higher in the vial in which the mutation was detected (12.9 vs. 11.6 and 30.4 vs. 23.6 ng/mL) but these differences were not statistically significant.
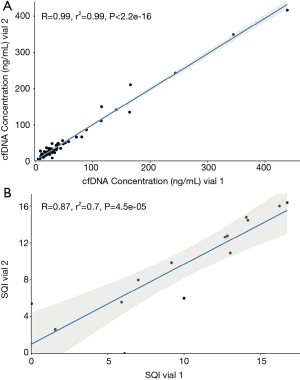
Among EGFR mutant cancer patients, the SQI values varied from 2.1 up to 25.0, indicating a large variability among patients. To further explore the SQI variability, we analyzed the SQI values based on the cfDNA concentration and the disease stage. The SQI value did not correlate with the cfDNA concentration (r2=0.17, P=0.28) or disease stage (stage III patients =9.1±3.1 and stage IV patients =11.5±4.8, P=0.41). We analyzed the SQI values based on the type of mutations, and found statistically significant differences between the SQI values for Ex19Del mutations (n=25 patients) and p.L858R (n=13 patients). The Ex19Del EGFR patients had higher SQI values compared to p.L858R EGFR patients (mean Ex19Del EGFR SQI =13.6 vs. mean p.L858R EGFR SQI =8.9; P=0.001) indicating that technical factors related to the detection of each mutation target impact the obtained SQI values.
Correlation between molecular status of cfDNA from plasma and tissue tumour
We evaluated the concordance between the molecular status in cfDNA and tissue biopsy in 60.1% (104/173) of patients (Table 4). The molecular analysis of the tumor was carried out by NGS in 93.3% (97/104) of patients, by the cobas® EGFR Test in tissue in 4.8% (5/104), by SCORPIONS and ARMS in 0.95% (1/104) and by 5’ NUCLEASE ASSAY in 0.95% (1/104). Activating EGFR mutations were detected in tissue tumor in 24% (25/104) of patients.
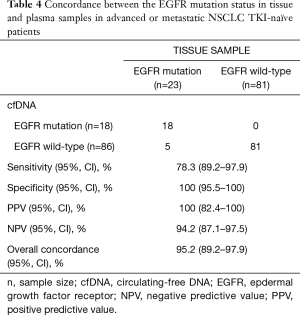
Full table
The overall accuracy of plasma cfDNA testing was 95.2%. Global sensitivity of EGFR testing in plasma was 78.3% and specificity 100%. The positive predictive value of the test was 100%, while the negative predictive value of the test was 94.2%.
Among patients with no EGFR mutations detected in cfDNA (n=86), seven of them harbored an EGFR mutation in the tumor tissue (8.2% of patients). Five discrepant cases were stage IV adenocarcinoma patients, one was a stage III adenocarcinoma patient and one was a stage IV NSCLC patient. Tissue testing identified an Ex19Del EGFR mutation in four tumors, a p.L858R in one tumor and an EGFR mutation not included in the cobas® EGFR Test in two tumors. In these two tumors, the alterations were a point mutation in exon 18 (c.211T>C; p.(Ile706Thr) plus a deletion in exon 19 in one case (c.2260_2277del18 (Lys754_Ile759del)) and an insertion in exon 20 (c.2317_2318insGTT) and an insertion in exon 20 (c.2317_2318insGTT) in the other case. Patients with concordant EGFR results in tissue and plasma and those with discordant results showed similar cfDNA concentration (28.8 vs. 30.3; P=0.73) suggesting that discordant results were not related to the initial amount of cfDNA. In addition, an oncogenic driver mutation in other genes was detected in 81.0% (64/79) of patients by tissue testing.
There was no available tumor tissue for molecular characterization in 39.9% (69/173) of NSCLC patients. Molecular characterization and posterior clinical management of these patients was carried out exclusively by testing the EGFR gene in plasma. We detected EGFR mutations in plasma in 34.8% of these cases (24/69), all of them diagnosed with adenocarcinoma, except for one patient diagnosed with NSCLC. Deletions of exon 19 and p.L858R point mutations accounted for 62.5% (15/24) and 33.3% (8/24) of cases respectively, while the G719X mutation was found in one case (4.2%).
Discussion
In the present study, we report the impact of introducing the analysis of cfDNA from plasma in a cohort of advanced or metastatic NSCLC TKI-naïve patients in a real-world clinical setting. The introduction of cfDNA testing in clinical diagnostics laboratories is a challenge for laboratory professionals (29) since both pre-analytical factors and the methodological approach used for cfDNA testing impact the robustness of the assay (30-32). The cobas® EGFR Test was introduced into the clinical practice of our hospital in 2017 to prospectively identify EGFR mutant NSCLC patients at baseline for treatment selection. In our study, the test showed a high diagnostic accuracy as previously reported in other studies (18,21). The clinical performance of the test reached similar values to other current approaches such as BEAMing dPCR (10) or NGS (20) which are more complex tests with longer turnaround times (8,33). In our cohort, we found EGFR mutations in 24.3% of patients and a mutation distribution similar to a recent study in NSCLC patients of European origin (21), but higher when compared to other cohorts (34,35). Such differences might be related to clinical and demographic characteristics, as well as patient selection biases. We found a distinct prevalence and distribution of EGFR mutations which depends on the histopathological tumor subtype and the clinical characteristics of the patients such as gender and smoking habit, as observed in NSCLC patient cohorts molecularly characterized by tumor tissue testing (33-35). This highlights the fact that cfDNA from plasma accurately reflects the molecular status of the tumor. A potential limitation of our study is that different approaches were used for tissue EGFR testing. However, NGS was used in the vast majority of patients, with various other methodologies used in the rest of the patients. All methods are adequate in terms of sensitivity and specificity (36,37) with the main difference being that NGS covers all possible mutations in the EGFR gene while the other methods are limited to specific EGFR mutations.
Although cfDNA is an optimal source for EGFR testing in NSCLC patients, undetectable EGFR mutations in plasma still are present in up to 20% of patients (18,35,38). In our cohort, a negative result by cfDNA testing was obtained in 8.2% of mutant EGFR NSCLC patients. Moreover, in two p.L858R EGFR NSCLC patients the test reported the mutation in one of the two distinct vials from the same blood extraction. The lack of detection of the EGFR mutation in cfDNA might be due to biological factors, such as a low concentration of cfDNA or low allelic fraction of ctDNA (18,39), or related to intrinsic technical factors of the assay. The analytical sensitivity of the Cobas EGFR Mutation Test v2 RT-PCR is limited to specific target EGFR mutations which have distinct sensitivity (14). Despite these limitations, implementing an initial EGFR testing in plasma prior to a tissue analysis in the clinical routine would accurately identify most of EGFR mutated locally in advanced or metastatic NSCLC patients with fast laboratory turnaround times. In addition, the straightforward analysis of cfDNA for EGFR testing would reduce the intrinsic issues related to the lack of tumor tissue for molecular characterization which occurs in a substantial subset of patients (8). This was supported by our cohort, where no tissue sample was available in 39.9% of patients. Among them, 34.8% of patients presented an EGFR mutation in cfDNA and received a targeted therapy immediately, underlining the important clinical impact of this strategy. Thus, the inclusion of EGFR analysis by non-invasive methods in the clinical setting is crucial to apply personalized medicine strategies especially in those NSCLC patients with no tissue tumor available. Although EGFR testing in plasma can reduce the time before starting treatment, a tissue biopsy is desirable when possible to properly classify patients histologically to avoid inappropriate treatment in patients with metastasis from an unknown primary, or primary lung cancer other than NSCLC, as TKIs are currently only approved for NSCLC.
In addition to an initial molecular stratification, cfDNA testing in NSCLC patients is mostly used for the evaluation of disease progression, prognosis, and treatment response. The mutant allele fraction (MAF) has been proposed as a potential marker for disease outcome in treated NSCLC patients since the MAF increases with the disease stage and tumor burden (40,41). Furthermore, a semi-quantitative index (SQI) of mutated allele fractional abundance (MAFA) has been correlated to a shorter progression-free survival and overall survival in KRAS mutated NSCLC patients (42). The cobas® EGFR Test provides a SQI value similar to the MAF value (8). We obtained significantly higher SQI values for the Ex19Del mutant tumors than for the L858R mutant tumors, and we did not observe any difference regarding disease stage or cfDNA concentration. In addition, we found no differences in cfDNA concentration among the EGFR mutations, as previously reported (43).
A pilot external quality assurance (EQA) study has previously demonstrated the imprecision of SQI for various EGFR mutations. The EQA study showed that distinct mutations with the same allele frequency resulted in different SQI values which were higher for the Ex19Del mutations (14). Moreover, the SQI value in our study showed only moderate reproducibility. A study conducted in samples from NSCLC patients observed that the SQI value does not accurately correlate with the number of mutant copies/mL (44). Thus, the lack of correlation between SQI and cfDNA concentration observed in our cohort suggest that technical factors impact the SQI value reported by the assay. This data indicates that the SQI value has only a moderate clinical utility as a follow-up biomarker in NSCLC patients since SQI variation during longitudinal monitoring of patients might not reflect changes in tumor development. Additional longitudinal studies in NSCLC patients are required to fully elucidate the relevance of the SQI value for each target mutation included in the assay prior to its implementation in the clinical routine.
The concentration of cfDNA has also been suggested as a plausible biomarker for tumor burden and prognosis in lung cancer patients (45-47). As observed in our cohort, higher levels of cfDNA are found in stage IV patients compared to stage III patients (48). We found a high diagnostic value of cfDNA concentration with a threshold value of 20 ng/mL to identify stage IV NSCLC patients with 72.3% sensitivity and 95% specificity. The obtained AUC value of 0.86 is excellent (49), indicating that cfDNA concentration is an optimal biomarker for diagnostic purposes. CfDNA levels are routinely measured, after cfDNA extraction, by accurate and standardized techniques (50). Thus, to include the cfDNA concentration result in the laboratory report would represent a fast and optimal strategy for NSCLC patient stratification.
The fragment pattern in NSCLC patients provides further information. Soliman et al. evaluated the diagnosis and prognosis value of cfDNA integrity by analyzing serum from NSCLC patients (46). They found that cfDNA integrity, calculated as the ratio between longer and shorter DNA fragments, is higher in NSCLC patients than in chronic obstructive pulmonary disease (COPD) patients and control subjects, and that lower cfDNA integrity is related with better survival. In our cohort, stage IV patients showed a higher fraction of 100–250 bp fragments. Apoptosis processes, which are associated with malignant progression and metastasis of cancer (51), promote the release of short cfDNA fragments. Thus, we hypothesized that the fraction of 100–250 bp fragments might be a biomarker for apoptosis in NSCLC patients. In fact, we found that the fraction of 100–250 bp showed a limited predictive value to identify stage IV NSCLC patients when compared to cfDNA concentration. To date, there is no consensus about the method used to measure cfDNA fragmentation. Current methods in use show variability in terms of the quality of the results (52). While capillary electrophoresis has shown good accuracy and reproducibility of the size distribution, it is sensitive to the presence of impurities in the samples, which could affect the results. However, it is simpler and faster than more accurate techniques such as quantitative real-time PCR (q-PCR), whose main disadvantage is the impact of gDNA contamination and requires the parallel analysis of reference samples; droplet digital PCR (ddPCR), which is sensitive to enzymatic inhibitors and might need a re-extraction process to ensure the quality of the sample; and NGS methods, which, while being the most accurate, are costly and time consuming. Additional studies are necessary to compare these more sensitive methods to capillary electrophoresis to assess the clinical significance of cfDNA fragmentation pattern as a biomarker in lung cancer patients and its potential introduction to clinical routine practice.
In summary, the introduction of plasma EGFR mutation analysis has proven useful as an initial approach for molecular characterization of locally advanced or metastatic NSCLC patients at diagnosis using a commercially available RT-PCR method. However, the low reproducibility and lack of correlation of the manufacturer’s SQI value with biological parameters limit its utility in the clinical practice. In addition, the study suggests that cfDNA concentration and cfDNA fragmentation pattern are interesting biomarkers for tumor stage in lung cancer patients.
Acknowledgments
The authors would like to dedicate this manuscript to Dr. Rafael Molina, who recently passed away and was a significant contributor to this study. His dedication and passion to the field of cancer were an inspiration to all of us. The authors also would like to thank the patients for their participation in the study and the study staff who were involved in collecting specimen material at the study sites. The authors would like to express our gratitude to Dr. James Hugall for providing language help for the manuscript.
Funding: This work was supported by Hoffman-La Roche Ltd., who supplied the reagents cobas® cfDNA Sample Preparation Kit and cobas® EGFR Mutation Test v2 to carry out the study. Hoffman-La Roche Ltd. has not been involved in the design, execution of the study, results interpretation nor manuscript elaboration.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-3142
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-3142
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-3142). Dr. NR reports personal fees from MSD, personal fees from AMGEM, personal fees from TAKEDA, personal fees from ASTRAZENECA, personal fees from NOVARTIS, personal fees from ABBVIE, personal fees from PFIZER, personal fees from GUARDANT HEALTH, personal fees from BOERINGHER INGELHEIM, personal fees from ROCHE, personal fees from BMS, outside the submitted work. Dr. NV reports personal fees from ROCHE, personal fees from PFIZER, personal fees from LILLY and other financial relationships with BMS, BOERINGHER INGELHEIM, LILLY and ASTRAZENECA, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The protocol was approved by the Hospital Clinic Ethics Committee (approval registration HCB/2016/0889) and was conducted in accordance with Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stewart BW, Wild CP. World Cancer Report 2014 [Online]. World Heal Organ. Epub ahead of print 2014. DOI: 9283204298.
- Inamura K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front Oncol 2017;7:193. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Nan X, Xie C, Yu X, et al. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 2017;8:75712-26. [Crossref] [PubMed]
- Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Patho. J Thorac Oncol 2013;8:823-59. [Crossref] [PubMed]
- Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-v27. [Crossref] [PubMed]
- Marchetti A, Palma JF, Felicioni L, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol 2015;10:1437-43. [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686). Clin Cancer Res 2016;22:2386-95. [Crossref] [PubMed]
- Arriola E, Paredes-Lario A, García-Gomez R, et al. Comparison of plasma ctDNA and tissue/cytology-based techniques for the detection of EGFR mutation status in advanced NSCLC: Spanish data subset from ASSESS. Clin Transl Oncol 2018;20:1261-7. [Crossref] [PubMed]
- Keppens C, Palma JF, Das PM, et al. Detection of EGFR Variants in Plasma: A Multilaboratory Comparison of a Real-Time PCR EGFR Mutation Test in Europe. J Mol Diagn 2018;20:483-94. [Crossref] [PubMed]
- Vollbrecht C, Lehmann A, Lenze D, et al. Validation and comparison of two NGS assays for the detection of EGFR T790M resistance mutation in liquid biopsies of NSCLC patients. Oncotarget 2018;9:18529-39. [Crossref] [PubMed]
- Kim Y, Shin S, Lee KA. A Comparative Study for Detection of EGFR Mutations in Plasma Cell-Free DNA in Korean Clinical Diagnostic Laboratories. Biomed Res Int 2018;2018:7392419 [Crossref] [PubMed]
- Singh AP, Li S, Cheng H. Circulating DNA in EGFR-mutated lung cancer. Ann Transl Med 2017;5:379. [Crossref] [PubMed]
- Normanno N, Denis MG, Thress KS, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017;8:12501-16. [Crossref] [PubMed]
- Szumera-Ciećkiewicz A, Olszewski WT, Tysarowski A, et al. EGFR mutation testing on cytological and histological samples in non-small cell lung cancer: A Polish, single institution study and systematic review of European incidence. Int J Clin Exp Pathol 2013;6:2800-12. [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Li Y, Xu H, Su S, et al. Clinical validation of a highly sensitive assay to detect EGFR mutations in plasma cell-free DNA from patients with advanced lung adenocarcinoma. PLoS One 2017;12:1-14. [Crossref] [PubMed]
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol 2019;30:597-603. [Crossref] [PubMed]
- Soria-Comes T, Palomar-Abril V, Ureste MM, et al. Real-World Data of the Correlation between EGFR Determination by Liquid Biopsy in Non-squamous Non-small Cell Lung Cancer (NSCLC) and the EGFR Profile in Tumor Biopsy. Pathol Oncol Res 2020;26:845-51. [Crossref] [PubMed]
- Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer 2015;113:476-83. [Crossref] [PubMed]
- Yoon KA, Park S, Lee SH, et al. Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls. J Mol Diagn 2009;11:182-5. [Crossref] [PubMed]
- Weng JL, Atyah M, Zhou CH, et al. Progress in quantitative technique of circulating cell free DNA and its role in cancer diagnosis and prognosis. Cancer Genet 2019;239:75-84. [Crossref] [PubMed]
- Provencio M, Torrente M, Calvo V, et al. Prognostic value of quantitative ctDNA levels in non small cell lung cancer patients. Oncotarget 2018;9:488-94. [Crossref] [PubMed]
- Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif 2019;17:100087 [Crossref] [PubMed]
- Lapin M, Oltedal S, Tjensvoll K, et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J Transl Med 2018;16:300. [Crossref] [PubMed]
- Arko-Boham B, Aryee NA, Blay RM, et al. Circulating cell-free DNA integrity as a diagnostic and prognostic marker for breast and prostate cancers. Cancer Genet 2019;235-236:65-71. [Crossref] [PubMed]
- Jiménez W. Liquid biopsy. A challenge for clinical laboratories. Adv Lab Med 2020;1.
- Arechederra M, Ávila MA, Berasain C. Liquid biopsy for cancer management: a revolutionary but still limited new tool for precision medicine. Adv Lab Med 2020;1.
- Bronkhorst AJ, Aucamp J, Pretorius PJ. Cell-free DNA: Preanalytical variables. Clin Chim Acta 2015;450:243-53. [Crossref] [PubMed]
- Sorber L, Zwaenepoel K, Jacobs J, et al. Circulating Cell-Free DNA and RNA Analysis as Liquid Biopsy: Optimal Centrifugation Protocol. Cancers (Basel) 2019;11:458. [Crossref] [PubMed]
- Steendam CMJ, Atmodimedjo P, de Jonge E, et al. Plasma Cell-Free DNA Testing of Patients With EGFR Mutant Non–Small-Cell Lung Cancer: Droplet Digital PCR Versus Next-Generation Sequencing Compared With Tissue-Based Results. JCO Precis Oncol 2019;1-9. [Crossref]
- Zhang YL, Yuan JQ, Wang KF, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016;7:78985-993. [Crossref] [PubMed]
- Reck M, Hagiwara K, Han B, et al. CtDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: The ASSESS study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Ellison G, Zhu G, Moulis A, et al. EGFR mutation testing in lung cancer: A review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol 2013;66:79-89. [Crossref] [PubMed]
- Mehta A, Vasudevan S, Sharma SK, et al. Biomarker testing for advanced lung cancer by next-generation sequencing; a valid method to achieve a comprehensive glimpse at mutational landscape. Appl Cancer Res 2020;40:4. [Crossref]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Alegre E, Fusco JP, Restituto P, et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumour Biol 2016;37:13687-94. [Crossref] [PubMed]
- Del Re M, Bordi P, Rofi E, et al. The amount of activating EGFR mutations in circulating cell-free DNA is a marker to monitor osimertinib response. Br J Cancer 2018;119:1252-8. [Crossref] [PubMed]
- Shen S, Wei Y, Zhang R, et al. Mutant-allele fraction heterogeneity is associated with non-small cell lung cancer patient survival. Oncol Lett 2018;15:795-802. [PubMed]
- Zulato E, Attili I, Pavan A, et al. Early assessment of KRAS mutation in cfDNA correlates with risk of progression and death in advanced non-small-cell lung cancer. Br J Cancer 2020;123:81-91. [Crossref] [PubMed]
- Zhang Y, Xu Y, Zhong W, et al. Total DNA input is a crucial determinant of the sensitivity of plasma cell-free DNA EGFR mutation detection using droplet digital PCR. Oncotarget 2017;8:5861-73. [Crossref] [PubMed]
- Macías M, Alegre E, Alkorta-Aranburu G, et al. The Dynamic Use of EGFR Mutation Analysis in Cell-Free DNA as a Follow-Up Biomarker during Different Treatment Lines in Non-Small-Cell Lung Cancer Patients. Dis Markers 2019;2019:7954921 [Crossref] [PubMed]
- Tissot C, Toffart AC, Villar S, et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J 2015;46:1773-80. [Crossref] [PubMed]
- Soliman SES, Alhanafy AM, Habib MSE, et al. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem Biophys Rep 2018;15:45-51. [Crossref] [PubMed]
- Hanibuchi M, Kanoh A, Kuramoto T, et al. Development, validation, and comparison of gene analysis methods for detecting EGFR mutation from non-small cell lung cancer patients-derived circulating free DNA. Oncotarget 2019;10:3654-666. [Crossref] [PubMed]
- Mirtavoos-Mahyari H, Ghafouri-Fard S, Khosravi A, et al. Circulating free DNA concentration as a marker of disease recurrence and metastatic potential in lung cancer. Clin Transl Med 2019;8:14. [Crossref] [PubMed]
- Mandrekar JN. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J Thorac Oncol 2010;5:1315-6. [Crossref] [PubMed]
- Ponti G, Maccaferri M, Manfredini M, et al. The value of fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of cell-free DNA (cfDNA) in malignant melanoma and prostate cancer patients. Clin Chim Acta 2018;479:14-9. [Crossref] [PubMed]
- Wang RA, Li QL, Li ZS, et al. Apoptosis drives cancer cells proliferate and metastasize. J Cell Mol Med 2013;17:205-11. [Crossref] [PubMed]
- Alcaide M, Cheung M, Hillman J, et al. Evaluating the quantity, quality and size distribution of cell-free DNA by multiplex droplet digital PCR. Sci Rep 2020;10:12564. [Crossref] [PubMed]


