
Prognostic nomograms and risk-stratifying systems for predicting survival in patients with resected pT2-4aN0M0 esophageal carcinoma
Introduction
Esophageal carcinoma (EC), the 8th most common cancer and 6th most common cause of cancer-related death worldwide, shows a gradual annual increase incidence (1-3). Although surgery has been considered the preferred first-line treatment for locally advanced EC (4), the survival rate differs substantially between cases. According to the latest version of National Comprehensive Cancer Network (NCCN) guidelines (5), follow-up is recommended for patients with completely resected pT2-4aN0M0 squamous cell carcinoma, and follow-up or adjuvant chemoradiation is recommended for patients with completely resected pT2-4aN0M0 adenocarcinoma. Regarding the variant prognosis of this population, the development of a prognostic model to facilitate the risk stratification and define high-risk cases that may benefit from closer follow-up or adjuvant therapy is imperative.
The 8th edition of the Tumor-Node-Metastasis (TNM) staging system of the American Joint Commission on Cancer (AJCC) is currently applied for predicting the survival of EC patients and guiding clinical practice (6). However, several previous studies have demonstrated the unsatisfactory predictive efficacy of the TNM staging system (7,8). Thus, additional prognostic factors to the TNM staging system, such as age, sex, chemotherapy information and lymph node harvested (LNH), should be considered when evaluated these patients.
In the current study, we analyzed resected cases of pT2-4aN0M0 EC from the Surveillance, Epidemiology, and End Results (SEER) database and Sun Yat-sen University Cancer Center (SYSUCC). The independent prognostic factors of overall survival (OS) and cancer specific survival (CSS) were identified, and prognostic nomograms were formulated. Risk-stratifying systems, based on the nomograms, were established to select the patients with an enhanced likelihood of poor survival. Different statistical methods were carried out to validate the models. Using the models, clinicians could refine treatment strategies and subsequently improve patient prognosis.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-3393).
Methods
Patient selection
Patients diagnosed with EC from 2004 to 2015 were retrospectively reviewed from the SEER database using SEER*Stat 8.3.5 software (https://seer.cancer.gov/seerstat/), and EC cases from 2001 to 2014 in the SYSUCC were also included. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent of enrolled patients and SYSUCC review board approval of were obtained in this study. The authenticity of the study has been validated by uploading the raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number RDDA2020001859.
The inclusion criteria were as follows: (I) pathologically diagnosed with squamous cell carcinoma or adenocarcinoma; (II) underwent surgery and (III) pT2-4aN0M0 (according to the 8th TNM staging system). The exclusion criteria were as follows: (I) age <18 years; (II) clinicopathological or follow-up information was unavailable and (III) perioperative death.
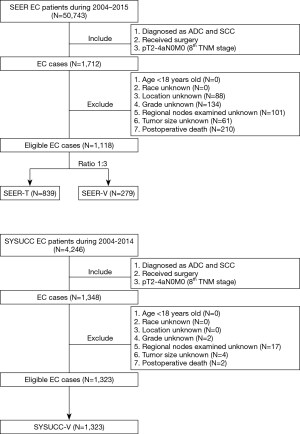
Figure 1 shows the detailed flowchart of this study. Cases with resected pT2-4aN0M0 EC from the SEER database were randomly divided into the training cohort (SEER-T) and the internal validation cohort (SEER-V) at a ratio of 3:1. Cases with pT2-4aN0M0 EC from the SYSUCC served as the external validation cohort (SYSUCC-V).
Data collection
Clinical and demographic variables including sex, age at diagnosis, surgical type, pTNM stage, histology, tumor grade, tumor size, LNH, tumor location, race, chemotherapy, radiotherapy, follow-up information, and vital status were retrospectively collected. Continuous variables such as age, LNH, and tumor size were transformed into categorical variables based on the cutoff values determined by the X-tile software (9). TNM staging was performed according to the 8th edition of the AJCC TNM staging system (6).
Statistical analysis
Statistical analysis was carried out by R version 3.5.2 (http://www.r-project.org), IBM SPSS Statistics (version 25.0; IBM, Armonk, NY, USA), X-tile software (9), and GraphPad Prism 8. OS was defined as the interval from the date of diagnosis to the date of death from any cause or the last follow-up. CSS was defined as the interval from the date of diagnosis to the date of lung cancer-related death or the last follow-up. All time events were estimated using the Kaplan-Meier method with a log-rank test. Multivariate analyses were applied to identify prognostic factors, and nomograms were developed. Each number/category of the independent prognostic variables in the nomogram was assigned a score on the point scale and finally added up to a total score. After that, a cutoff value, achieved by X-tile software, was applied to transform the continuous score into2 categories (low-risk and high-risk groups). The concordance index (C-index) (10), calibration curves and decision curve analysis (DCA) (11) were applied to verify the predicted effect of the nomogram. Pearson’s χ2 test or Fisher’s exact test was used to compare categorical variables between groups. Two-sided P<0.05 was considered statistically significant.
Results
Patients characteristics
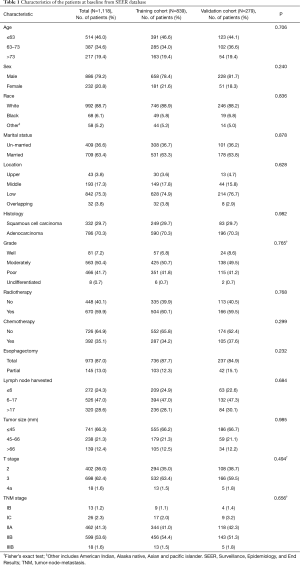
Between 2004 and 2015, a total of 50,743 EC cases from the SEER database were reviewed. The aforementioned selection criteria yielded a study population of 1,118 eligible EC patients (SEER-T, n=839 cases; SEER-V, n=279 cases). The general clinical features are listed in Table 1. The median age was 65 years old (range, 26–91 years). Almost all the patients were white (88.7%). Male patients (79.2%) and adenocarcinoma (70.3%) accounted for a substantial proportion of the entire cohort. There was a slightly high percentage of low-third tumors (75.3%). Over half of patients received radiotherapy (59.9%). The median follow-up time was 30.0 months (range, 1.0–155.0 months). All covariates between the training cohort and the validation cohort were well-balanced (Table 1). The clinical characteristics of the SYSUCC-V cohort are summarized in Table S1.
Full table
Cox regression analysis
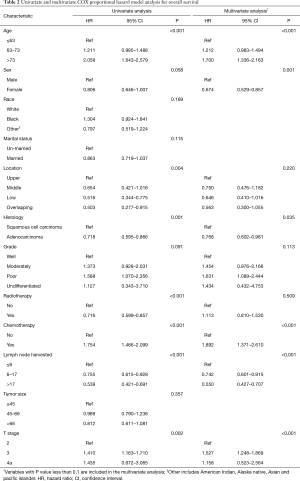
Regarding OS, a univariate Cox regression analysis suggested that age, sex, location, histology, tumor grade, radiotherapy, chemotherapy, LNH, and T stage were associated with patient’s survival (Table 2). The multivariate analysis confirmed that age, sex, histology, chemotherapy, LNH, and T stage were independent factors that impacted OS (Table 2).
Full table
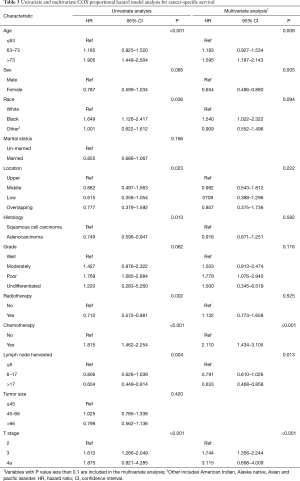
As for CSS, the univariate Cox regression analysis revealed that age, sex, race, location, histology, tumor grade, radiotherapy, chemotherapy, LNH, and T stage were prognostic predictors (Table 3). The multivariate analysis confirmed that age, sex, chemotherapy, LNH, and T stage were statistically significant prognostic factors (Table 3).
Full table
Nomograms and risk-stratifying Systems
Statistically significant factors from the multivariate analysis were entered into the nomogram, which was developed to calculate 3, 5 and 10-year OS and CSS probabilities (Figure 2). With regard to OS, the nomogram indicated that LNH was the strongest predicator, followed by age and chemotherapy (Figure 2A). Concerning CSS, the graphic demonstrated that T stage was the strongest predicator, followed by LNH and age (Figure 2B).
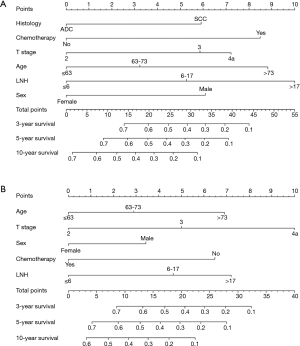
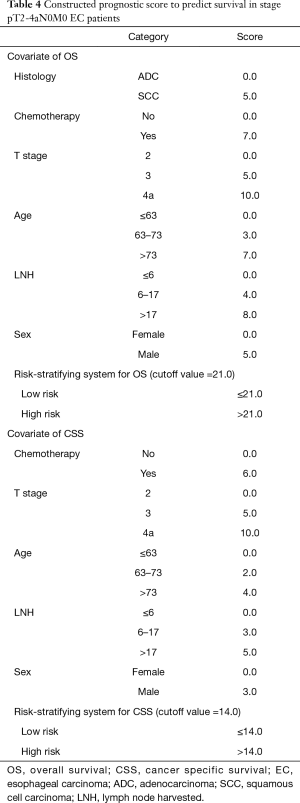
In further analyses, each independent prognostic variable was assigned a risk score according to the point scale using R software. A total score was calculated. The total cutoff values (21.0 for OS and 14.0 for CSS), determined by X-tile software, were used to dichotomize EC cases into low-risk and high-risk subgroups (Table 4).
Full table
Calibration and validation
For OS, the C-index was 0.69 [95% confidence interval (CI): 0.66–0.72], 0.64 (95% CI: 0.59–0.69), and 0.62 (95% CI: 0.61–0.63) for the SEER-T cohort, the SEER-V cohort, and the SYSUCC-V cohort, respectively. For CSS, the C-index was 0.68 (95% CI: 0.66–0.71), 0.67 (95% CI: 0.63–0.71), and 0.61 (95% CI: 0.59–0.63) for the SEER-T, SEER-V, and SYSUCC-V, respectively. The calibration curves indicated an optimal consistency between predicted and actual observations (Figure S1 for OS and Figure S2 for CSS). The DCA demonstrated that the nomograms had a satisfying clinical net benefit when compared with the TNM staging system (Figure S3A for OS and Figure S3B for CSS).
Stratification efficacy evaluation
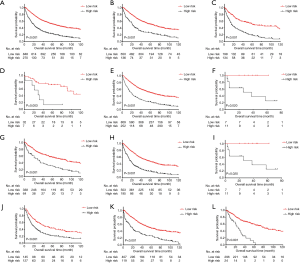
The total score cutoff values were used to dichotomize EC cases into low-risk and high-risk subgroups. Kaplan-Meier curves for OS in the entire cohort, stratified by histology, TNM stage, T stage, and LNH were used to evaluate the risk-stratifying systems’ efficacy. The low-risk subgroup’s OS was significantly better than the high-risk subgroup for the entire cohort (P<0.001, Figure 3A). For histology, both in the squamous cell carcinoma group and adenocarcinoma group, low-risk patients had significantly favorable OS than high-risk patients (adenocarcinoma, P<0.001, Figure 3B; squamous cell carcinoma, P<0.001, Figure 3C). In the TNM stage subset analysis, low-risk cases had high survival levels compared with high-risk cases in all the three substages (stage I, P=0.003, Figure 3D; stage II, P<0.001, Figure 3E; stage IIIB, P=0.020, Figure 3F). In the T stage subsets analysis, low-risk cases had better OS rate compared with high-risk cases in all the three T substages (T2, P<0.001, Figure 3G; T3, P<0.001, Figure 3H; T4a, P=0.020, Figure 3I). Regarding LNH, the OS of low-risk cases was also superior than that of high-risk cases in all the three categories (LNH ≤6, P<0.001, Figure 3J; 6< LNH ≤17, P<0.001, Figure 3K; LNH >17, P<0.001, Figure 3L).
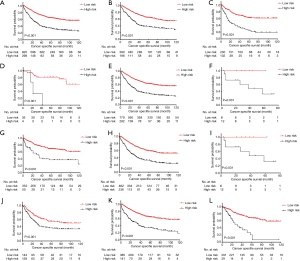
As for CSS, the survival curves of these two different risk subgroups, in the full analysis set (P<0.001, Figure 4A) or stratified by histology(adenocarcinoma, P<0.001, Figure 4B; squamous cell carcinoma, P<0.001, Figure 4C), TNM stage (stage I, P<0.001, Figure 4D; stage II, P<0.001, Figure 4E; stage IIIB, P=0.031, Figure 4F), T stage (T2, P<0.001, Figure 4G; T3, P<0.001, Figure 4H; T4a, P=0.031, Figure 4I) and LNH categories (LNH ≤6, P=0.001, Figure 4J; 6< LNH ≤17, P<0.001, Figure 4K; LNH > 17, P<0.001, Figure 4L), also showed significant distinctions.
Discussion
In the present study, novel prognostic nomograms, including routinely available factors such as age, sex, histology, chemotherapy, LNH, and T stage, was established to predict OS and CSS. The risk-stratifying systems stratified patients with resected pT2-4aN0M0 EC into two different prognostic groups. The validation of the models using the C-index, DCA, and calibration curves demonstrated its good performances. With the help of the easy-to-use models, clinicians could identify a subset of EC patients with poor survival who may need closer follow-up or intervention therapy.
According to the NCCN guidelines for EC (5), surveillance is recommended for patients with completely resected pT2-4aN0M0 squamous cell carcinoma, and surveillance or adjuvant chemoradiation is recommended for patients with completely resected pT2-4aN0M0 adenocarcinoma. Previous studies have suggested a dismal 5-year survival rate for resectable EC (12). Therefore, it was necessary to identify the prognostic factors of resected pT2-4AN0M0 EC and formulate a nomogram to determine high-risk patients who may be the best adjuvant therapy candidates.
Compared with advanced tumors, the clinical manifestation of relapse or progression of nodal negative tumors might be less obvious. As we all know, there are still many developing countries in the world. Esophageal carcinoma mostly occurs in the group of people with relatively poor economic situation, and most of them live in rural area far away. The expensive follow-up examinations and long distances form major obstacles for these patients to get scheduled follow-up examinations. Based on our classifying systems, we strongly recommend that the high-risk patients should return to hospital for a scheduled follow-up examination or even postoperative therapy, irrespective of cost.
According to the current 8th edition of the AJCC TNM staging system (6), the covariates adopted to subdivide pT2-4aN0M0 EC are depth of tumor invasion, location, grade, and histology. Our nomograms confirmed that the TNM stage factors such as depth of tumor invasion and histology were the independent prognostic predictors, but grade and location were not. From our perspective, the relatively small number of cases might interpret, to a certain extent, the no statistically significance of grade. As is widely acknowledged, patients with upper-third EC are unlikely to undergo surgery due to anatomical difficulties and technique challenges (4), and the limited treatment options may impair the survival of these patients. Only 43 (3.8%) upper-third EC patients in our cohort, and all cases included in our study underwent surgical resection. Therefore, the impact of location on long-term survival was not deep enough to reach statistically significance. In our nomograms, 4 other prognostic factors (age, sex, LNH, and chemotherapy) were also included, which contributed to the nomograms’ superior predictive value compared with the TNM staging system.
Despite several previously reported predictive models (7,8,13-20), a nomogram for resected pT2-4aN0M0 EC from two large databases with long-term follow-up has not been established. In the study by Du et al. (15), the authors reviewed data of 4,566 resected localized EC from the SEER database and developed a predictive nomogram. Unfortunately, there was unavailable clinicopathological information in their research, which could affect the model's predictive efficacy. Wu et al. (16) also analyzed 20,623 EC adenocarcinoma cases from the SEER dataset and established a nomogram to predict OS and CSS. Grade, T stage, N stage, M stage, underwent surgery, insurance record and marital status were entered into their nomogram which showed great performance (C index: 0.720–0.733) when compared with our nomogram. However, chemotherapy, a critical prognostic factor for survival, was lacking in their study. Moreover, the tumor stage was diagnosed either pathologically or clinically due to the inclusion criteria in their research, which may lead to bias. Molecular indicators such as integrated mRNA-lncRNA signature (21), Linc-PINT (22) and progranulin (23) were also reported in several prognostic nomograms. In our study however, only routinely available variables such as sex, age, histology, chemotherapy, T stage and LNH were used to formulate the nomogram, making the nomogram convenient in clinical practice. Furthermore, the nomogram was externally validated by another large cohort, and the results were generally applicable. Large case numbers with long-term follow-up and the use of both internal and external validation make our nomogram reliable.
Our study also has several limitations. First, the factors used to formulate the nomograms represented only a portion of clinicopathological characteristics owing to the SEER database, which had limited relevant information. Second, the quality of several data in the SEER database is relatively poor, for example, the median number of lymph node harvested in the SEER database is only 12, which is less than the number (at least 15 examined lymph nodes) recommended by the NCCN guideline (24), and it might lead to stage migration. Third, although the developed prognostic score models showed good discrimination capacity, further large-scale validation cohorts are warranted. Finally, the retrospective nature of the study may have contributed to selection bias. Further efforts on prospective study results collection and broader prognostic factors recruitment such as the timing and regimen of chemotherapy, timing and dosage of radiotherapy, resection margin, tumor markers, and genetic molecule indicators are encouraged to improve this nomogram.
In conclusion, the novel nomograms demonstrated good prognostic prediction for patients with resected pT2-4aN0M0 EC and stratified the population subset into low-risk and high-risk prognostic subgroups, which may aid individual clinical practice.
Acknowledgments
We thank AME Editing Service for language assistance.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-3393
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-3393
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-3393). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Uzunoglu FG, Reeh M, Kutup A, et al. Surgery of esophageal cancer. Langenbecks Arch Surg 2013;398:189-93. [Crossref] [PubMed]
- Milleron B, Westeel V, Gounant V, et al. Pathological complete response: A predictive survival factor after neoadjuvant chemotherapy in lung cancer. Bull Cancer 2016;103:66-72. [Crossref] [PubMed]
- Rice TW, Gress DM, Patil DT, et al. Cancer of the esophagus and esophagogastric junction-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:304-17.
- Zheng Y, Fu S, He T, et al. Predicting prognosis in resected esophageal squamous cell carcinoma using a clinical nomogram and recursive partitioning analysis. Eur J Surg Oncol 2018;44:1199-204. [Crossref] [PubMed]
- Su D, Zhou X, Chen Q, et al. Prognostic Nomogram for Thoracic Esophageal Squamous Cell Carcinoma after Radical Esophagectomy. Plos One 2015;10:e0124437 [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Harrell FE Jr, Califf RM, Pryor DB, et al. Evaluating the yield of medical tests. JAMA 1982;247:2543-6. [Crossref] [PubMed]
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. [Crossref] [PubMed]
- Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023-75. [Crossref] [PubMed]
- Zhu C, You Y, Liu S, et al. A Nomogram to Predict Distant Metastasis for Patients with Esophageal Cancer. Oncol Res Treat 2020;43:2-9. [Crossref] [PubMed]
- Min BH, Yang JW, Min YW, et al. Nomogram for prediction of lymph node metastasis in patients with superficial esophageal squamous cell carcinoma. J Gastroenterol Hepatol 2020;35:1009-15. [Crossref] [PubMed]
- Du F, Sun Z, Jia J, et al. Development and Validation of an Individualized Nomogram for Predicting Survival in Patients with Esophageal Carcinoma after Resection. J Cancer 2020;11:4023-9. [Crossref] [PubMed]
- Wu XX, Chen RP, Chen RC, et al. Nomogram predicting cancer-specific mortality in patients with esophageal adenocarcinoma: a competing risk analysis. J Thorac Dis 2019;11:2990-3003. [Crossref] [PubMed]
- Tang X, Zhou X, Li Y, et al. A Novel Nomogram and Risk Classification System Predicting the Cancer-Specific Survival of Patients with Initially Diagnosed Metastatic Esophageal Cancer: A SEER-Based Study. Ann Surg Oncol 2019;26:321-8. [Crossref] [PubMed]
- Deng W, Zhang W, Yang J, et al. Nomogram to Predict Overall Survival for Thoracic Esophageal Squamous Cell Carcinoma Patients After Radical Esophagectomy. Ann Surg Oncol 2019;26:2890-8. [Crossref] [PubMed]
- Zhang WY, Chen XX, Chen WH, et al. Nomograms for predicting risk of locoregional recurrence and distant metastases for esophageal cancer patients after radical esophagectomy. BMC Cancer 2018;18:879. [Crossref] [PubMed]
- Xie K, Liu S, Liu J. Nomogram predicts survival benefit for non- metastatic esophageal cancer patients who underwent preoperative radiotherapy. Cancer Manag Res 2018;10:3657-68. [Crossref] [PubMed]
- Lan T, Xiao Z, Luo H, et al. Bioinformatics analysis of esophageal cancer unveils an integrated mRNA-lncRNA signature for predicting prognosis. Oncol Lett 2020;19:1434-42. [PubMed]
- Zhang L, Chen J, Wang L, et al. Linc-PINT acted as a tumor suppressor by sponging miR-543 and miR-576-5p in esophageal cancer. J Cell Biochem 2019;120:19345-57. [Crossref] [PubMed]
- Li G, Dong T, Yang D, et al. Progranulin promotes lymphangiogenesis through VEGF-C and is an independent risk factor in human esophageal cancers. Hum Pathol 2018;75:116-24. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Esophageal and Esophagogastric Junction Cancers. Version 1.2019. Available online: https://www.nccn.org/professionals/physician_gls/pdf/esophageal.pdf.


