
Effects of miR-132-3p on progress and epithelial mesenchymal transition of non-small cell lung cancer via regulating KLF7
Introduction
Non-small cell lung cancer (NSCLC) possesses higher incidence rate (1). The occurrence of NSCLC is a multi-step process, involving cumulative genetic and epigenetic changes (2,3). Nevertheless, pathogenesis mechanisms relating to NSCLC have not yet been fully elucidated.
MicroRNAs (miRNAs) are endogenous non-coding RNA with an average of 22 nucleotides, and exert post-transcriptive influence among animals and plants (4). The pro- and anti-cancer genes regulated by miRNAs extensively take part in metastasis and progression of NSCLC (5). Research has found that 37 miRNAs are abnormally expressed in lung cancer, and 24 of them are related to NSCLC (6,7). Nevertheless, the role and mechanism of miR-132-3p in NSCLC remain unclear.
This research aimed to elucidate the roles of miR-132-3p and KLF7 in the progression of NSCLC, in order to yield practical direction for NSCLC treatment. In this study, the authors first found that miR-132-3p can affect NSCLC proliferation by targeting KLF7 expression, invasion, migration and EMT process.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-353).
Methods
Clinical specimens
A total of 30 pairs of NSCLC and peripheral normal tissue specimens were obtained from Shanghai Tongren Hospital. Age: 31–65. Sex: (male, 19 cases; female, 11 cases). The samples were stored at −80 °C for standby. All procedures involving human participants in this study are consistent with the Helsinki Declaration (as revised in 2013). The study has got approval from Ethics Committee of Shanghai Tongren Hospital (No. 2018-067-01) and obtained the informed consent forms of all patients.
Cell culture
Normal human pulmonary epithelial cell lines (BEAS-2B, GNHu27)and five kinds of NSCLC cell lines (A549, TCHu150) (H1944, SCSP-596) (H292, TCHu122) (H1299, TCHU160) (H1650, TCHu152) were obtained from the cell bank of Chinese Academy of Sciences (Shanghai, China). Roswell Park Memorial Institute 1640 (RPMI-1640) medium with 10% fetal bovine serum (FBS) was applied to cultivate cellular systems.
Cell transfection
The miR-132-3p inhibitor was infected with A549 and H1944 severally. Separately, miR-132-3p was infected with H1650 and H1299, individually, to determine luciferase activity. In another experiment, A549 was infected with LV-antimiR-132-3p and LV-antitimiR-132-3p + shKLF7. Also, H1650 was infected with LV-miR-132-3p, LVmiR-132-3p + LV-KLF7, and LV-NC. The role of miR-132-3p and KLF7 on the expression of the abovementioned cells was measured.
Quantitative RT-PCR analysis
Trizol reagents were used to harvest total RNAs from target tissues and cells. According to manufacturer directions, we adopted deoxyribonuclease I (DNase I) to cleave DNA in the experiment. The specific primers were as follows: miR-132-3p (5'-TCC ACC AAG AAG CTG AGC GAG-3'; 5'-GTC CAG CCC ATG ATG GTT CTG TC-3'), KLF7 (5'-AGA CAT GCC TTG AAT TGG AAC G-3'; 5'-GTC CAG CCC ATG ATG GTT CT-3'). The primers for miR-132-3p and KLF7 were obtained from Novland Bio-pharmaceutical Technology Co., Ltd (Shanghai, China).
Luciferase analysis
Firstly, the WT 3’-NCR sequence as well as MT 3’-NCR sequence were cloned into psiCHECK-2 luciferase reporter vector Shanghai Beinuo Biotechnology Co., Ltd. (Shanghai, China). Then, the cell line 293T was infected with luciferase reporter vector (Shanghai Bitai Biotechnology Co., Ltd., Shanghai, China). The activity of luciferase was detected by dual luciferase reporter system (Guangzhou Huijun Biotechnology Co., Ltd, Guangdong, China).
Western blot
Cells were obtained 2 days after transfection. After quantifying through bicinchoninic (BCA), the total protein was set in lanes with 15 µg each. After blocking with fat-free milk, primary antibodies KLF7 (13125-1-AP, Proteintech), E-cadherin (20874-1-AP, Proteintech), vimentin (ab92547, Abcam), as well as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (60004-1-Ig, Proteintech) were incubated at 4 °C for 1 night. Then, the secondary antibodies were incubated together with the membranes.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded in a 96-well plate (1×104). Later, CCK-8 was added to the wells. The optical density (OD) value was measured under 450 nm wavelength using an Epoch reader (Shanghai Woyuan Technology Co., Ltd., Shanghai, China).
Transwell assay
A 100 µL quantity of cellular suspension and 200 µL of serum-free culture fluid were mixed into the Transwell upper chamber. Then, the complete culture medium with 400 µL 0.05% FBS were incubated together in the Transwell lower chamber for 2 days with 5% CO2. Cells that migrated under membrane surfaces were fixed and dried and dyed with crystal violet. The average field was chosen under microscope.
Edu experiment
The cells were mixed with 5-Ethynyl-2’deoxyuridine (Edu) solution, fixed with 4% paraformaldehyde, 0.5% Triton X-100 (Shanghai Tongwei Biology Co., Ltd) was used for penetration enhancement, and they were then stained with Click-iT reagent (Thermo Fisher, Waltham, MA, USA). Pictures were taken under microscope (Shanghai Yuguang Instrument Co., Ltd.).
Statistical analysis
Data was described as and was analyzed by t-test between different groups. All experimental organisms were repeated 3 times. A P value <0.05 indicated a significant difference.
Results
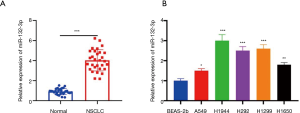
More miR-132-3p was expressed in NSCLC
Firstly, the expression of miR-132-3p in NSCLC was determined. The outcomes revealed its expression was elevated in 5 cell lines (Figure 1A,B).
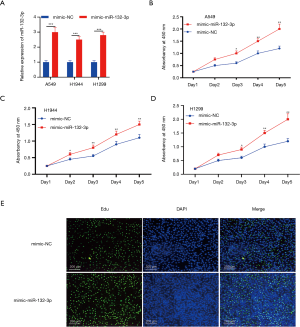
High miR-132-3p expression boosted NSCLC proliferation
It was revealed that miR-132-3p expression in A549, H1944, and H1299 was raised via miR-132-3p mimic transfection (Figure 2A). Correspondingly, the CCK-8 outcomes displayed the same trend (Figure 2B,C,D). The miR-132-3p mimic accelerated the growth of positive lung cancer cells (Figure 2E).
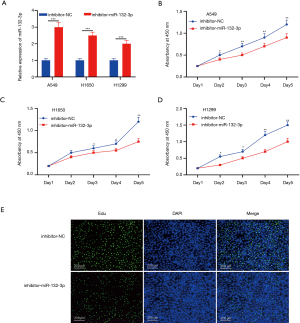
miR-132-3p low-expression controlled NSCLC proliferation
Low-expression of miR-132-3p was exhibited in its inhibitor transfection (detailed in Figure 3A). More surprisingly, the survival rate of A549, H1650, and H1299 cells lessened dramatically (Figure 3B,C,D). The EdU staining implied that the positive lung cancer cells were substantially lessened when exposed to miR-132-3p inhibitor (Figure 3E).
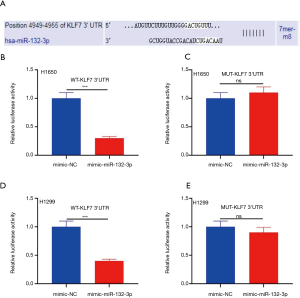
KLF7 was a downstream target of miR-132-3p
Our findings supported that KLF7 is a downstream target of miR-132-3p (Figure 4A). The activity of mimic miR-132-3p was substantially lessened in H1650 via WT-KLF7 3'UTR transfection (Figure 4B). Curiously, in H1650, MUT-KLF7 3'UTR transfection did not cause any changes (Figure 4C). The activity of mimic miR-132-3p in H1299 was same as in H1650 (Figure 4D). Similarly, In H1299, MUT-KLF7 3'UTR transfection did not lead to any changes (Figure 4E).
miR-132-3p negatively regulated KLF7
As shown in Figure 5A,B, low expression of KLF7 was exhibited via shKLF7 transfection. The expression of KLF7 was increased via A549 cells transfected with LV-anti-miR-132-3p (Figure 5C,D). Yet, LV-KLF7 transfection displayed the opposite trend (Figure 5E,F). Additionally, KLF7 expression in H1650 displayed the opposite trend (Figure 5G,H).
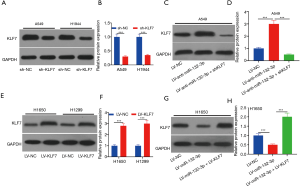
miR-132-3p promoted cell development via adjusting KLF7
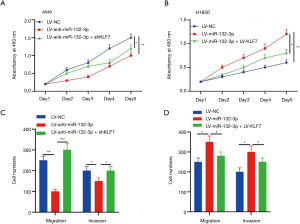
The outcomes of CCK-8 revealed that the cell vitality in A549 appeared substantially lower in LV-anti-miR-132-3p + shKLF7 (Figure 6A). The cell viability of LV-miR-132-3p was higher than LV-miR-132-3p + LV-KLF7 (Figure 6B). Transwell assay showed that the number of cells decreased in LV-anti-miR-132-3p + shKLF7 (Figure 6C). Meanwhile, Transwell results indicated that the number of cells raised in LV-miR-132-3p (Figure 6D).
The miR-132-3p/KLF7 axis adjusted interstitial protein
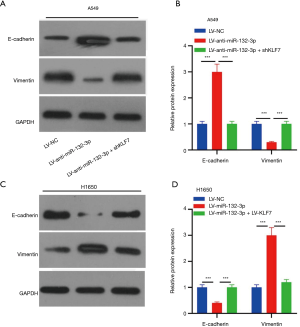
As displayed in Figure 7A,B, the expression of in cells infected with LV-anti-miR-132-3p + shKLF7 was greatly reduced, while the expression of vimentin exhibited an opposite trend (Figure 7A,B). Surprisingly, LV-miR-132-3p + LV-KLF7 transfection not only increased E-cadherin expression but also reduced the expression of vimentin (Figure 7C,D).
Discussion
The tumor that is NSCLC is complex and malignant (8). The pathogenic factors of NSCLC include environment, diet, infection, host genes, and so on. Abnormal gene expression is the key factor of NSCLC, especially the abnormal expression of oncogenes or tumor suppressor genes (9-16). Abnormal expression of miR-132-3p has been confirmed in glioma, breast cancer, and bladder cancer (17-24). However, the role of miR-132-3p in the progression of NSCLC has remained unclear.
In this study, we discovered more miR-132-3p in tissue specimens and cells. A negative relationship was detected between KLF7 and miR-132-3p. In addition, miR-132-3p mimic facilitated the development of NSCLC tumor. Nevertheless, the inhibitor of miR-132-3p exhibited a contrasting result. The above-mentioned results hinted that miR-132-3p performed an oncogenic role, while KLF7 exerted an anti-oncogenic role in NSCLC.
EMT was accompanied by changes in tumor cytomorphology, adhesion between cells and between cells and stroma lost, but the invasion and migration existed, which promoted tumor metastasis in the body (25). Numerous studies have shown that the differentiated miRNA expression in tumors is highly related to the occurrence of tumor EMT (26,27). Our research investigated E-cadherin and vimentin, the markers of EMT, to explore any possible latent mechanisms. It has been shown that miR-132-3p controls EMT and inactivates the PI3K/Akt pathway in NSCLC. In addition, Li et al. have found that regulating KLF7 can inhibit NSCLC invasion, migration, and EMT (28). In the current study, miR-132-3p/KLF7 was shown to be involved in the progression of NSCLC via adjusting relevant EMT expression.
At present, the treatment measures for NSCLC mainly include surgical excision and chemotherapy, however, its survival rate is poor, especially the chemical resistance of cancer cells seriously reducing the survival rate of NSCLC patients (29). In recent years, the search for new treatment methods has become a hot topic of research. To be sure, studies of targets such as exosomes, FOXO1 signaling pathway, miRNAs, PI3K/Akt signaling pathway, Wnt signaling pathway, LncRNAs, NF-κB signaling pathway are possible for in-depth understanding and treatment of NSCLC (30). In the future, more clinically viable drugs will need to be developed for these targets.
In conclusion, miR-132-3p could directly target KLF7 expression and regulate EMT transformation, thus accelerating the migration and invasion of NSCLC cells. This provided strong data support for a better understanding of miR-132-3p regulating NSCLC development. More importantly, the miR-132-3p/KLF7 axis presents a new clinical approach for the diagnosis, treatment, and prevention of NSCLC metastasis.
Acknowledgments
Funding: This work was supported by Changning District Health and Family Planning Commission (No. 20164Y004) and Changning District Medical Specialty (Chest disease diagnosis and treatment specialty, No. YXMZK008).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-353
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-353
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-353). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures involving human participants in this study are consistent with the Helsinki Declaration (as revised in 2013). The study has got approval from Ethics Committee of Shanghai Tongren Hospital (No. 2018-067-01) and obtained the informed consent forms of all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Aoi T. Biology of lung cancer: genetic mutation, epithelial-mesenchymal transition, and cancer stem cells. Gen Thorac Cardiovasc Surg 2016;64:517-23. [Crossref] [PubMed]
- Dong N, Shi L, Wang DC, et al. Role of epigenetics in lung cancer heterogeneity and clinical implication. Semin Cell Dev Biol 2017;64:18-25. [Crossref] [PubMed]
- Kang SM, Lee HJ. MicroRNAs in human lung cancer. Exp Biol Med (Maywood) 2014;239:1505-13. [Crossref] [PubMed]
- Slaby O, Svoboda M, Michalek J, et al. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer 2009;8:102. [Crossref] [PubMed]
- Zhang WC, Liu J, Xu X, et al. The role of microRNAs in lung cancer progression. Med Oncol 2013;30:675. [Crossref] [PubMed]
- Del Vescovo V, Grasso M, Barbareschi M, et al. MicroRNAs as lung cancer biomarkers. World J Clin Oncol 2014;5:604-20. [Crossref] [PubMed]
- Yin Q, Fischer L, Noethling C, et al. In vitro-assessment of putative antiprogestin activities of phytochemicals and synthetic UV absorbers in human endometrial Ishikawa cells. Gynecol Endocrinol 2015;31:578-81. [Crossref] [PubMed]
- Chen W, Zheng R, Zeng H, et al. Annual report on status of cancer in China, 2011. Chin J Cancer Res 2015;27:2-12. [Crossref] [PubMed]
- Cortinovis D, Monica V, Pietrantonio F, et al. MicroRNAs in non-small cell lung cancer: current status and future therapeutic promises. Curr Pharm Des 2014;20:3982-90. [Crossref] [PubMed]
- Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J 2012;18:268-74. [Crossref] [PubMed]
- Vannini I, Fanini F, Fabbri M. MicroRNAs as lung cancer biomarkers and key players in lung carcinogenesis. Clin Biochem 2013;46:918-25. [Crossref] [PubMed]
- Tsang FH, Au SL, Wei L, et al. MicroRNA-142-3p and microRNA-142-5p are downregulated in hepatocellular carcinoma and exhibit synergistic effects on cell motility. Front Med 2015;9:331-43. [Crossref] [PubMed]
- Chang F, Lee JT, Navolanic PM, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia 2003;17:590-603. [Crossref] [PubMed]
- Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis 2004;9:667-76. [Crossref] [PubMed]
- Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal 2002;14:381-95. [Crossref] [PubMed]
- Zhang M, Li Y, Wang H, et al. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther 2019;20:524-36. [Crossref] [PubMed]
- Gu Y, Cai R, Zhang C, et al. miR-132-3p boosts caveolae-mediated transcellular transport in glioma endothelial cells by targeting PTEN/PI3K/PKB/Src/Cav-1 signaling pathway. FASEB J 2019;33:441-54. [Crossref] [PubMed]
- Li S, Xu JJ, Zhang QY. MicroRNA-132-3p inhibits tumor malignant progression by regulating lysosomal-associated protein transmembrane 4 beta in breast cancer. Cancer Sci 2019;110:3098-109. [Crossref] [PubMed]
- Liu P, Li X, Guo X, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif 2019;52:e12614 [Crossref] [PubMed]
- Arab A, Karimipoor M, Irani S, et al. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet 2017;216-217:150-8. [Crossref] [PubMed]
- Cai Y, Wang W, Guo H, et al. miR-9-5p, miR-124-3p, and miR-132-3p regulate BCL2L11 in tuberous sclerosis complex angiomyolipoma. Lab Invest 2018;98:856-70. [Crossref] [PubMed]
- Zhang F, Lin X, Yang X, et al. MicroRNA-132-3p suppresses type I IFN response through targeting IRF1 to facilitate H1N1 influenza A virus infection. Biosci Rep 2019;39:BSR20192769 [Crossref] [PubMed]
- Xia L, Li D, Lin C, et al. Comparative study of joint bioinformatics analysis of underlying potential of 'neurimmiR', miR-212-3P/miR-132-3P, being involved in epilepsy and its emerging role in human cancer. Oncotarget 2017;8:40668-82. [Crossref] [PubMed]
- Aggarwal V, Montoya CA, Donnenberg VS, et al. Interplay between tumor microenvironment and partial EMT as the driver of tumor progression. iScience 2021;24:102113 [Crossref] [PubMed]
- Du G, Yu X, Chen Y, et al. MiR-1-3p Suppresses Colorectal Cancer Cell Proliferation and Metastasis by Inhibiting YWHAZ-Mediated Epithelial-Mesenchymal Transition. Front Oncol 2021;11:634596 [Crossref] [PubMed]
- Pan G, Liu Y, Shang L, et al. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond) 2021;41:199-217. [Crossref] [PubMed]
- Li K, Yuan C. MicroRNA-103 modulates tumor progression by targeting KLF7 in non-small cell lung cancer. Int J Mol Med 2020;46:1013-28. [Crossref] [PubMed]
- Dziedzic R, Marjański T, Rzyman W. A narrative review of invasive diagnostics and treatment of early lung cancer. Transl Lung Cancer Res 2021;10:1110-23. [Crossref] [PubMed]
- Ashrafizadeh M, Zarrabi A, Hushmandi K, et al. Association of the Epithelial-Mesenchymal Transition (EMT) with Cisplatin Resistance. Int J Mol Sci 2020;21:4002. [Crossref] [PubMed]
(English Language Editor: J. Jones)


