
MicroRNA-155-5p promotes cell proliferation and invasion in lung squamous cell carcinoma through negative regulation of fibroblast growth factor 9 expression
Introduction
Lung cancer (LC) claims more lives than any other malignancy, with the International Organization for Cancer Risk Research estimating that approximately 1.59 million people die due to LC each year worldwide (1,2). Non-small cell lung cancer (NSCLC) accounts for the largest proportion of LC diagnoses, comprising 80–85% of all cases (3). Although the treatment effect for some subgroups of patients with NSCLC has been significantly improved by great breakthroughs in treatment technology, including surgery, radiotherapy, chemotherapy, and molecular targeted therapies such as epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) tyrosine kinase inhibitors, the overall survival (OS) rate after 5 years is still only 11% (4). Therefore, the discovery of novel targets that are of great significance in the occurrence and development of NSCLC is urgently needed.
MicroRNAs (miRNAs, miRs) are small non-protein coding RNAs that are vital participants in many biological processes (5,6). MiRNAs usually work by base pairing to the 3’ untranslated region (3’-UTR) of their target genes (5). Research has revealed miRNAs as critical players in cellular processes including proliferation, apoptosis, autophagy, migration, and invasion (7). Further, miRNAs are known to be abnormally expressed during the occurrence and progression of LC, esophageal cancer, neuroblastoma, and lymphocytic leukemia (5). miR-155 shows strong sequence homology in humans, mice, and chickens in the exons of non-coding RNA transcribed from the B cell integration cluster (BIC) found on chromosome 21. It has a conservative function in evolution and is expressed in lymphoid organs (8,9). miR-155-5p has key involvement in the innate and adaptive immune response, and proinflammatory mediators (interleukin 1 beta, tumor necrosis factor-alpha, and Toll-like receptors) and monocytes/macrophages can stimulate its expression (10,11). It is related to different types of cancer, cardiovascular diseases, and viral infections. Of the miRNAs discovered to date, miR-155 is among those most frequently implicated in malignancies. High miR-155 expression is closely related to drug-resistance and poor prognosis in a variety of tumors (12,13).
Our previous studies have shown that miR-155 can promote the proliferative and invasive abilities of NSCLC via its target programmed cell death protein 4 (14). In lung squamous cell carcinoma (SCC) (15), which is a major subtype of NSCLC, Wang et al. revealed miR-372-3p to promote the growth and metastatic activity of cells by targeting fibroblast growth factor 9 (FGF9). Still, the precise relevance of the miR-155-FGF9 relationship to the pathogenesis and development of lung SCC remains unclear. Based on previous studies, we speculated that miR-155 is expressed at higher levels, while FGF9 is expressed at lower levels, in lung SCC tissues in comparison with normal tissues, and that miR-155 acts as a regulator in lung SCC occurrence and development via targeting the FGF9 gene. This study aimed to explore the relationship between microRNA-155-5p (miR-155-5p) and FGF9 gene expression and their effects on the proliferation and invasion of lung SCC through experiments. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-882).
Methods
Collection of clinical samples
Forty pairs of surgically resected lung SCC tissues and paracancerous tissues (>5 cm from the edge of the cancer bed tissue) from patients who were treated in our hospital between August 2017 and February 2019 were collected. The tissues were stored in liquid nitrogen. The patients included 20 men and 20 women, ranging in age from 55-75 years old. All patients were treatment-naïve before their operations. Immediately after resection, phosphate-buffered saline (PBS) was used to rinse the tissues, which were then stored in liquid nitrogen for subsequent RNA and protein extraction. A signed informed consent form was obtained from each patient. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Approval for this research was granted by the ethics committee of the Nanjing Lishui People’s Hospital (NO: 201769518).
Shanghai Cell Bank of Chinese Academy of Sciences supplied the lung SCC cell lines (H2170, H1703, and H520) and the normal lung bronchial epithelial cell line BEAS-2B used in our experiments. Cell Counting Kit-8 (CCK-8), crystal violet, and a protein quantitative kit were bought from Beijing Biyuntian Biotechnology Co., Ltd (China), and kits to extract total RNA and perform reverse transcription-polymerase chain reaction (RT-PCR) were obtained from Takara (Japan). The primary antibody Anti-β-actin, FGF9, and horseradish peroxidase labeled secondary antibody were purchased from CST company. Roswell Park Memorial Institute 1640 (RPMI-1640) Medium, Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), and double antibody were supplied by Gibco (USA). An RT-PCR instrument (American ABI Company, BJ001271) and an inverted microscope (Japan Nikon Company, ECLIPSE MA100) were also purchased.
RT-PCR detection of miR-155-5p and FGF9 expression
The total RNA extraction kit was used to extract total RNA from the all tissues and cell lines in strict accordance with the manufacturer’s instructions. The RT-PCR kit was used to obtain complementary DNA. RT-PCR was performed with SYBR Green dye. We applied the ∆∆Ct method for calculation of the relative expressions of miR-155-5p and FGF9, with U6 RNA and β-actin serving as the internal controls, respectively.
Vector construction and transfection
We obtained miR-155-5p and NC inhibitors, and miR-155-5p and NC mimics from Shanghai Jimma Biotechnology Co., Ltd. The amplified sequence of the 3'-UTR of FGF9 messenger RNA (mRNA) was:
- Forward: 5'-TGTACAGGCCAGATAGGCATTTT-3';
- Reverse: 5'-GAAACATGTACAGGCCA-3'.
The amplified product was cloned into the pGL3 vector, and the FGF9 overexpression vector was also supplied by Shanghai Jikai Gene Co., Ltd. Lipofectamine 2000 was used for transfection of the cell lines, which were then screened using G418 to ensure stable expression and subjected to subsequent functional experiments.
Western blot detection
Tissue and cell proteins were collected and lysed on ice. The protein concentrations were determined through bicinchoninic acid (BCA) protein quantification. Sample amounts of 50 µg were then added to the wells individually. Proteins were electrophoresized on a 12% polyacrylamide gel, under 90V and 120V with continuous pressure. After completion, the proteins were transferred to a polyvinylidene difluoride membrane, PBS was added, and the membrane was subjected to sealing with 5% skim milk powder at room temperature for 2 hours. After the addition of primary antibodies, the membrane was incubated overnight at 4 °C, with β-actin used as the internal reference protein. Following that, the membrane was incubated with a peroxidase-labeled secondary antibody (1:2,000) at room temperature for 2 hours. Finally, it was observed in a gel imager after color development.
CCK-8 assay
For the detection of cell viability, we performed a CCK-8 assay. First, transfected logarithmic-growth-phase H2170 cells were collected and diluted with DMEM supplemented with 10% FBS to a density of 5×104/mL. The suspended cells were then inoculated onto a 96-well culture plate, with 200 µL per well, and incubated in a cell culture incubator under 37 °C, 5% CO2. There were 5 parallel wells in each cell group. CCK-8 reagent (10 µL) was added at days 1, 2, 3, 4, and 5. After 3-hour incubation at 37 °C in an incubator, cell viability was detected at 490 nm using a full-wavelength microplate reader, and the optical density value was obtained. We performed this experiment in triplicate. The absorbance values of 5 replicate wells were averaged, and the cell growth curves were plotted.
Clone formation assay
Following digestion with 0.25% trypsin, logarithmic-growth-phase cells were pipetted into a single cell suspension. After centrifugation at 1,000 rpm at room temperature for 5 minutes, the supernatant was discarded. The cell suspension was diluted in multiples of gradient, inoculated into a petri dish at an appropriate density, rotated gently to disperse the cells evenly, and then cultivated in a carbon dioxide incubator at 37 °C, 5% CO2. The medium was changed at 2- to 3-day intervals, and cell growth was observed regularly. When macroscopic colonies formed, after approximately 2–3 weeks, the medium was discarded, and the cells were carefully washed with PBS twice and fixed with 4% paraformaldehyde for 15 minutes. Then, the fixing solution was discarded, and the cells were twice washed with PBS buffer. The cells were then stained with crystal violet solution, following which, they were subjected to thorough rinsing under running water 3 times. Finally, images were taken; and the clone-forming rate was analyzed. This experiment was repeated 3 times.
Transwell assay
To examine the invasion and migration abilities of cells, we performed a Transwell assay. First, a pipette was used to remove and discard the old medium in the culture plate. New serum-free medium was added to each well, and the cells were left at room temperature for 30 minutes. Then, the upper surface of the Transwell’s lower chamber was coated with 50 mg/L Matrigel (diluted to a ratio of 1:8), and placed at 37 °C for half an hour to allow the Matrigel to polymerize into a gel. Cells in the logarithmic growth phase were collected and washed 1–2 times with PBS buffer. The cells were then digested with trypsin before the digestion was stopped and the cells were centrifuged to remove the culture medium. Serum-free medium containing BSA was used to resuspend the cells, with the cell density adjusted to 5×105 Pcs/mL. Cell suspension (100 µL) was added to the upper chamber, and 600 µL of culture medium containing 20% FBS was added to the bottom compartment. Bubbles can occur between the lower culture medium and the cells; if there are bubbles, the chemotaxis of the lower culture medium will be weakened or even eliminated. If bubbles appeared, the chamber was lifted, removed, and put in the culture plate. Three replicates were made for each well. The Transwells were placed in a constant temperature incubator at 37 °C and 5% CO2 for 48 hours. After that, the Transwell chambers were taken out, and the medium in the wells was discarded. The cells were washed 2–3 times with PBS buffer, and then fixed with 600 µL of methanol for half an hour. Following that, a pipette was used to remove the culture medium, and the cells were washed 2–3 times with PBS buffer. Finally, the cells were stained with crystal violet for 10 minutes and washed 3 times with PBS. Five fields of view were randomly selected, and the cells were observed and counted under a 100× microscope. The average value was calculated based on the results of 3 replicate wells.
In vivo experiment
Sixteen female BALB/c nu/nu thymus-deficient nude mice (4–6 weeks old; weighing 18–20 g) were obtained from Beijing Vitong Lihua Laboratory Animal Technology Co., Ltd (China). All of the mice were disinfected after purchase and bred adaptively in an indoor culture environment for 1 week, before being reared with 4 mice in each cage in a specific-pathogen-free environment. The mice were housed under a 12-hour/12-hour light/dark cycle, with relative humidity (50%±10%) and temperature (22±2) °C, and regular ultraviolet irradiation for disinfection. The mice could drink freely and were given sterilized special mouse pellet feed. Experiments were performed under a project license (No.: 201786292) granted by the Nanjing Lishui People’s Hospital, in compliance with Chinese guidelines for the care and use of animals.
To establish the mouse model, transfected cells in a good growing state were trypsinized, and washed 2–3 times with PBS buffer. A single cell suspension was then made and centrifuged at 1,000 rpm for 5 minutes. After the removal of the supernatant, the cells were resuspended in PBS buffer, the cells were counted, and finally, the cell density was adjusted to 1×107 cells/ml. Four nude mice were chosen from each group and their backs were disinfected with iodophor. Following that, cell suspension (200 µL) was drawn using a disposable sterile syringe, and gently shaken to mix the cells evenly. Then, the cell suspension was injected into the mice via their backs. Under the same conditions described above, the feeding, drinking and activity status of the mice were regularly observed every day for a 30-day period; during this time, the weights and tumor sizes of the mice were monitored. After 30 days, cervical dislocation was performed to euthanize the mice. Finally, the tumors were extracted, and the wet weight was weighed.
Statistical analysis
Data were analyzed using SPSS20.0 and presented as means ± standard deviations (SDs). Groups were compared through t-tests or analysis of variance, with P<0.05 taken to indicate statistical significance.
Results
miR-155-5p and FGF9 levels are negatively correlated in lung SCC tissue
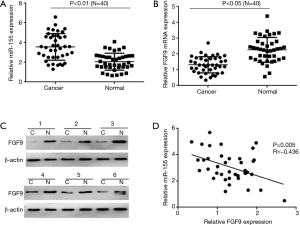
Tissue samples from 40 patients with lung SCC were subjected to fluorescence quantitative PCR to determine the levels of miR-155-5p and FGF9. We found that in lung SCC tissue in comparison to adjacent tissue, miR-155-5p was highly expressed (Figure 1A), whereas FGF9 was under-expressed (Figure 1B). We also carried out western blot to detect FGF9 expression in tissue samples from 6 patients and found that the results were consistent with those of fluorescence quantitative PCR (Figure 1C). Correlation analysis of miR-155-5p and FGF9 expression in SCC tissue uncovered a significant negative correlation between the 2 (Figure 1D). The above results show that miR-155-5p and FGF9 have critical participation in SCC development.
miR-155-5p targets and negatively regulates the expression of FGF9
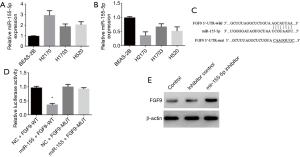
To further study the specific functions of FGF9 and miR-155-5p in lung SCC cells, we performed fluorescence quantitative PCR to detect miR-155-5p and FGF9 expression in lung SCC lines (H2170, H1703, and H520) and a normal bronchial epithelial cell line (BEAS-2B). The results showed that H2170 cells exhibited the highest expression of miR-155-5p and the lowest expression of FGF9 (Figure 2A,B); consequently, the H2170 cell line was used for subsequent functional experiments. Using bioinformatics software to predict the relationship between FGF9 and miR-155-5p, we found that miR-155-5p may bind to the 3’-UTR region of FGF9 to regulate its expression (Figure 2C). To further determine the effect of 155-5p on FGF9 expression, a dual luciferase reporter gene plasmid was constructed, and the binding sequence of miR-155 and FGF9 was inserted into the 3’-UTR of the plasmid, with the binding sequence mutation sequence used as a negative control (Figure 2D). miR-155 mimic was transfected into the H2170 cell line, and after 24 hours, the cell lysate was collected. The results revealed miR-155-5p overexpression resulted in significant inhibition of wild-type (WT) FGF9 activity, but had no significant effect on the mutant (Mut) FGF9. To further observe how miR-155-5p affects FGF9 protein expression, the H2170 cell line was subjected to transfection with miR-155-5p inhibitor and inhibitor control, and western blot was subsequently performed to detect FGF9 expression (Figure 2E). We observed that miR-155-5p inhibition resulted in significant elevation of FGF9 expression. The above experiments indicate that FGF9 could be targeted to attain downstream regulation of miR-155-5p in H2170 cells.
miR-155-5p inhibits the proliferative and cloning abilities of lung SCC cells by inhibiting FGF9 expression
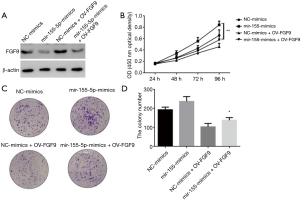
Next, we further studied whether miR-155-5p exerts its biological functions by inhibiting the expression of FGF9, by transfecting H2170 cells with miR-155-5p mimics, control mimics, miR-155-5p mimics + Ov-FGF9, or control mimics + Ov-FGF9 (Figure 3). CCK-8 and plate clone formation assays were carried out to detect cell proliferation ability. First, the western blot results showed that compared with the control mimics group, overexpression of miR-155-5p significantly inhibited FGF9 expression. In the control mimics + Ov-FGF9 group, FGF9 expression was significantly promoted, but overexpression of miR-155-5p could significantly reverse the upregulation of FGF9 (Figure 3A,B). The CCK-8 test results showed that in comparison with the control mimics group, miR-155-5p overexpression significantly promoted SCC cell proliferation, while overexpression of FGF9 resulted in significant inhibition of SCC cells’ proliferative ability. Overexpression of FGF9 in cells could significantly reverse the cell proliferation promoted by miR-155-5p. The clone formation experiment also showed that overexpression of FGF9 could significantly reverse the cell clone formation ability promoted by miR-155-5p (Figure 3C,D). The above results indicate that miR-155-5p exerts an inhibitive effect on the proliferative and cloning abilities of lung SCC cells via inhibiting the expression of FGF9.
miR-155-5p exerts inhibitory effects on lung SCC cell invasion and migration via inhibiting FGF9 expression
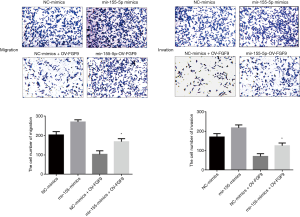
To further study whether miR-155-5p exerts its biological functions by inhibiting the expression of FGF9, we transfected H2170 cells with miR-155-5p mimics, control mimics, miR-155-5p mimics + Ov-FGF9, or control mimics + Ov-FGF9, and examined their invasion and migration abilities by performing Transwell experiments (Figure 4). We found that compared with the control mimics group, miR-155-5p overexpression could significantly promote the ability of SCC cells to invade and migrate; meanwhile, FGF9 overexpression (control mimics+ Ov-FGF9) produced the opposite effects, and it could significantly inhibit the promoting effects of miR-155-5p mimics on cell invasion and migration.
Overexpression of FGF9 can significantly block the tumorigenic activity of miR-155-5p
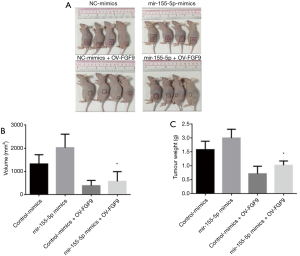
We further verified the regulatory relationship between miR-155-5p and FGF9 expression by transfecting H2170 cells with miR-155-5p mimics, control mimics, miR-155-5p mimics + Ov-FGF9, or control mimics + Ov-FGF9. A total of 106 cells were collected. Cells from each group were injected into the skin of nude mice for 6-8 weeks (4 in each group). We observed the tumor growth, and recorded their sizes and shapes daily. The mice were sacrificed 4 weeks after the injection, and the tumor tissues were extracted. The results show that in comparison with the control mimics group, miR-155-5p overexpression significantly promoted SCC cell proliferation in vivo; meanwhile, overexpression of FGF9 (Control mimics + Ov-FGF9) produced the opposite effect, and resulted in significant inhibition of the promoting effect of miR-155-5p mimics on SCC cell proliferation in vivo (Figure 5A,B). The tumor-bearing tissue was weighed, and FGF9 overexpression was found to result in significant inhibition of the promoting effect of miR-155-5p mimics on SCC cell proliferation in vivo (Figure 5C). These observations indicate that overexpressing FGF9 can block the ability of miR-155-5p to promote SCC cell proliferation in vivo.
Discussion
LC is associated with high rates of morbidity and mortality globally. Although the treatment, early diagnostic methods, and technological detection of LC have all seen great progress in recent years, this disease still ranks among the top contributors to tumor-associated deaths (1). Therefore, exploring the molecular regulatory mechanisms of LC occurrence and development and elucidating the cancer-promoting effects of key signaling molecules is critical to developing targeted drugs and early diagnostic technologies for patients with LC. This study is the first to prove that the expression of miR-155-5p in lung SCC cells and tissue is negatively correlated with that of FGF9. Our in vitro and in vivo functional experiments revealed miR-155-5p to be an oncogene whose negative regulation of FGF9 expression promotes the proliferation and invasion of lung SCC.
Recently, numerous researchers have reported that related genes being vital players in LC occurrence and development (6). Research shows that miR-148a inhibits the migratory and invasive potential of cells through targeting and regulating Wnt1 expression (16). Moreover, miRNAs can also serve as early diagnostic molecular markers in LC, as well as therapeutic targets and treatment evaluation markers (6,17). miR-30a expression in plasma is significantly elevated in patients with early LC, so its detection may be a new non-invasive method for early screening and differential diagnosis of patients with NSCLC (18). In LC, diminished miRNA-21 expression can also serve as a measurement of the treatment effect. For instance, for patients with advanced stages of the disease, Kanglaite injection can significantly reduce the expression of miRNA-21 in the serum (19). Here, we found that miR-155-5p and FGF9 are expressed at a high and low level, respectively, in lung SCC tissues; further, correlation analysis showed that the 2 have a negative correlation. We further used bioinformatics software to predict the downstream target of miR-155-5p, which showed that FGF9 may be a target. The dual-luciferase reporter assay results confirmed that FGF9 is the downstream regulatory target of miR-155-5p.
FGF9 is one of the members of the fibroblast growth factor family (20). Research has uncovered the abnormal expression of this gene in multiple tumor types, including LC, prostate cancer, and colon cancer, suggesting that it may be of vital importance to tumor occurrence and development (21-23). Also, Wang et al. found FGF9 expression to be up-regulated in patients with lung adenocarcinoma, with its abnormal expression potentially inhibiting disease progression (15). The present work found that FGF9 showed a significant expression reduction in lung SCC tissues and cells, while its overexpression significantly inhibited cell proliferation, clone formation, and invasion; these findings are consistent with those of other research. In lung SCC, miR-372-3p promotes the growth and metastatic abilities of cells through its downregulation of FGF9 (15). However, some studies have shown that FGF9 plays a role in promoting LC. For instance, one study found that miR-4317 inhibited NSCLC proliferation and invasion through its targeting of FGF9 (24), while another reported that miR-187 targeted FGF9 to inhibit cervical cancer cell growth (21). The above results indicate that FGF9 has 2-sided effects on tumors, and different upstream regulatory molecules may determine whether it has a tumor-promoting or anti-tumor function.
miR-155-5p has pivotal involvement in the regulation of a variety of tumors. For instance, in liver cancer, it acts as a promoter of cell proliferation and an inhibitor of cell apoptosis through its direct targeting of the 3'-UTR of gene of phosphate and tension homology deleted on chromsome ten (PTEN). Molecular mechanism studies have shown that miR-155-5p promotes liver cancer by targeting PTEN and activating the PI3K/Akt pathway (25). In addition, significant upregulation of miR-155-5p has been observed in cervical cancer tissues compared with non-cancerous tissues. In cervical cancer, downregulating the expression of miR-155-5p inhibits the ability of cervical cancer cells to grow, migrate, and invade in vitro, and miR-155-5p regulates disease development by regulating TP53INP1 expression (26).
The present study has uncovered miR-155-5p to be a new regulatory molecule that regulates FGF9 in lung SCC. Functional studies have shown that inhibiting the expression of miR-155-5p can significantly promote the expression of FGF9, while overexpression of miR-155-5p can significantly inhibit the expression of FGF9. Our cell function experiments also proved that overexpression of FGF9 can significantly block the promoting effects of miR-155-5p on cancer cell proliferation and invasion. Also, subcutaneous tumor formation experiments in nude mice further evidenced miR-155-5p’s role as a promoter of lung SCC occurrence and development through its inhibition of FGF9 expression.
In conclusion, this study is the first to prove that lung SCC tissues and cells exhibit a high expression of miR-155-5p and a low expression of FGF9, with the 2 displaying a negative correlation. Our molecular mechanism experiments showed that miR-155-5p acts as a promoter of lung SCC cell proliferation and invasion through its targeting and negative regulation of FGF9. In vivo experiments further proved that FGF9 overexpression can block the cancer-promoting effects of miR-155-5p. This study has uncovered the relationship between miR155-5p and FGF9 in lung SCC, and thus, has potentially identified novel molecular markers or targets for early diagnosis or targeted therapy for patients with this disease.
Acknowledgments
Funding: Lishui District People’s Hospital Project (No.: 2019-02).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-882
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-882
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-882). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. A signed informed consent form was obtained from each patient. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Approval for this research was granted by the ethics committee of the Nanjing Lishui People’s Hospital (No: 201769518). Experiments were performed under a project license (No.: 201786292) granted by the Nanjing Lishui People’s Hospital, in compliance with Chinese guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat Res 2016;170:25-46. [Crossref] [PubMed]
- Romaszko AM, Doboszyńska A. Multiple primary lung cancer: A literature review. Adv Clin Exp Med 2018;27:725-30. [Crossref] [PubMed]
- Nasim F, Moua T. Lung cancer in combined pulmonary fibrosis and emphysema: a large retrospective cohort analysis. ERJ Open Res 2020;6:00521-2020 [Crossref] [PubMed]
- Oberndorfer F, Müllauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol 2018;30:69-76. [Crossref] [PubMed]
- Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol 2017;1509:1-10. [Crossref] [PubMed]
- Wu M, Wang G, Tian W, et al. MiRNA-based Therapeutics for Lung Cancer. Curr Pharm Des 2018;23:5989-96. [Crossref] [PubMed]
- Van Roosbroeck K, Calin GA. Cancer Hallmarks and MicroRNAs: The Therapeutic Connection. Adv Cancer Res 2017;135:119-49. [Crossref] [PubMed]
- Mahesh G, Biswas R. MicroRNA-155: A Master Regulator of Inflammation. J Interferon Cytokine Res 2019;39:321-30. [Crossref] [PubMed]
- Zhou H, Li J, Gao P, et al. miR-155: A Novel Target in Allergic Asthma. Int J Mol Sci 2016;17:1773. [Crossref] [PubMed]
- Bala S, Csak T, Saha B, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol 2016;64:1378-87. [Crossref] [PubMed]
- Zhang X, Ji W, Huang R, et al. MicroRNA-155 is a potential molecular marker of natural killer/T-cell lymphoma. Oncotarget 2016;7:53808-19. [Crossref] [PubMed]
- Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev 2018;37:33-44. [Crossref] [PubMed]
- Khoshinani HM, Afshar S, Pashaki AS, et al. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Jpn J Radiol 2017;35:664-72. [Crossref] [PubMed]
- Liu F, Song D, Wu Y, et al. MiR-155 inhibits proliferation and invasion by directly targeting PDCD4 in non-small cell lung cancer. Thorac Cancer 2017;8:613-9. [Crossref] [PubMed]
- Wang Q, Liu S, Zhao X, et al. MiR-372-3p promotes cell growth and metastasis by targeting FGF9 in lung squamous cell carcinoma. Cancer Med 2017;6:1323-30. [Crossref] [PubMed]
- Chen Y, Min L, Ren C, et al. miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One 2017;12:e0171751 [Crossref] [PubMed]
- Du X, Zhang J, Wang J, et al. Role of miRNA in Lung Cancer-Potential Biomarkers and Therapies. Curr Pharm Des 2018;23:5997-6010. [Crossref] [PubMed]
- Sun L, Chen Y, Su Q, et al. Increased Plasma miRNA-30a as a Biomarker for Non-Small Cell Lung Cancer. Med Sci Monit 2016;22:647-55. [Crossref] [PubMed]
- Wu Y, Zhang J, Hong Y, et al. Effects of Kanglaite Injection on Serum miRNA-21 in Patients with Advanced Lung Cancer. Med Sci Monit 2018;24:2901-6. [Crossref] [PubMed]
- Cui G, Shao M, Gu X, et al. The value of FGF9 as a novel biomarker in the diagnosis of prostate cancer. Artif Cells Nanomed Biotechnol 2019;47:2241-5. [Crossref] [PubMed]
- Liang H, Luo R, Chen X, et al. miR-187 inhibits the growth of cervical cancer cells by targeting FGF9. Oncol Rep 2017;38:1977-84. [Crossref] [PubMed]
- Mizukami T, Togashi Y, Naruki S, et al. Significance of FGF9 gene in resistance to anti-EGFR therapies targeting colorectal cancer: A subset of colorectal cancer patients with FGF9 upregulation may be resistant to anti-EGFR therapies. Mol Carcinog 2017;56:106-17. [Crossref] [PubMed]
- Siegfried JM, Farooqui M, Rothenberger NJ, et al. Interaction between the estrogen receptor and fibroblast growth factor receptor pathways in non-small cell lung cancer. Oncotarget 2017;8:24063-76. [Crossref] [PubMed]
- He X, Chen SY, Yang Z, et al. miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J Exp Clin Cancer Res 2018;37:230. [Crossref] [PubMed]
- Fu X, Wen H, Jing L, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci 2017;108:620-31. [Crossref] [PubMed]
- Li N, Cui T, Guo W, et al. MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther 2019;12:3181-96. [Crossref] [PubMed]


