
This article has an erratum available at: http://dx.doi.org/10.21037/jtd-2021-38 the article has been update on 2021-09-01 at here.
Clinicopathological models for predicting lymph node metastasis in patients with early-stage lung adenocarcinoma: the application of machine learning algorithms
Introduction
Lung cancer has been reported to be the most common cancer type worldwide and the leading cause of cancer death (1). Among lung cancer cases that have various pathological characteristics, 80–85% of them can be categorized as non-small cell lung cancer (NSCLC) (2). In the treatment of NSCLC, lymph node dissection (LND) during radical surgery is considered crucial (3). A better understanding of lymph node metastasis (LNM) pattern aids to demarcate the extent of LND. Many studies focused on LNM in late-stage lung cancer, while LNM in small-size NSCLC should not be ignored as it could have an incidence rate up to 10% (4,5). Moreover, occult LNM (OLNM) occurred not rarely in early-stage NSCLC (6-8), which might lead to a poor prognosis, especially for patients who received sublobar resection and sublevel excision of lymph nodes. Thus, it is more than necessary to precisely evaluate the risk of LNM intraoperatively and postoperatively, even in patients with no preoperatively suspected involvement of lymph nodes.
Machine learning (ML) generally defines an algorithm-based process that predicts outcome from large data files, presuming the existence of a pattern amidst the data that will identify the outcome. Comparing to traditional statistical models, ML predictive analysis has several benefits, including less outcomes required for each predictor, no requirement for specific hypothesis and allowance of interaction between variables (9,10). ML-based predictive analysis has been validly used in medical field (11,12). From the authors’ perspective, there were very few studies that have reported the application of ML algorithms for evaluating the risk of LNM in lung cancer patients. This study aims to find validated ML models for the prediction of LNM in early-stage adenocarcinomas incorporating the clinical characteristics and postoperative histological patterns.
We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-98).
Methods
Study population
This study enrolled 709 NSCLC patients who has received lobectomy with systematic LND at Peking Union Medical College Hospital from January 2013 to December 2019. Enrolled patients had single foci NSCLC with maximum diameter ≤2 cm on CT. Patients who met any one of the following conditions were excluded: (I) diagnosed with small cell lung cancer; (II) diagnosed with multiple lung cancer; (III) preoperative radiation therapy or chemotherapy; (IV) distant metastasis; (V) incomplete clinical information. All enrolled patients received lobectomy plus systematic LND. The scope of systematic LND included N1 nodes (#10, #11, #12, #13, and #14), 2R, 4R, 3A, 3P, #7, #8, and #9 for tumors located in the right lung, and 4L, #5, #6, #7, #8, and #9 for tumors located in the left lung. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Peking Union Medical College Hospital (No. S-K1049) and informed consent was taken from all the patients.
Clinicopathological characteristics
This study enrolled a total of 19 variables in three categories. Preoperative clinical characteristics included age, gender, smoking status, and serum carcinoembryonic antigen (CEA). Radiographical features were recorded from CT by one radiologist and two thoracic clinicians independently, which included tumor imaging density, tumor side, tumor maximum diameter and specific signs as spiculation, vessel convergence, lobulation and pleural indentation. Disagreement was solved by their consensus. According to postsurgical histology, cancer lesions were divided into four subtypes, atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), microinvasive adenocarcinoma (MIA) and invasive adenocarcinoma (IA) (13). AAH, a precancerous condition, was included into this study as it had a lot in common with early-stage lung cancer. For all tumor lesions, histological details were further examined by pathological experts at our hospital, which included the presence of papillary, micropapillary, solid, acinar and lepidic components. Additionally, lymphovascular invasion (LVI) and pleural invasion (PI) were also considered risk factors for LNM. Pathological staging was based on 8th edition TNM Classification for lung cancer (14). PET-CT examination results were not analyzed because patients were not regularly recommended to receive the expensive PET-CT examination which was not covered by national medical insurance yet.
Development and validation of ML-based models
Firstly, z-score normalization was preprocessed to code running for continuous variables except for multinomial Naïve Bayes (MNB) algorithm, to which min-max normalization was done (15). For the prediction of LNM, we applied two conventional models including logistic regression (LR) and MNB, and six representative supervised ML algorithms including adaptive boosting (ADB), artificial neural network (ANN), decision tree (DT), gradient boosting decision tree (GBDT), random forest classifier (RFC) and extreme gradient boosting (XGB) (16-21).
Overfitting, meaning model becomes too specific to be suitable for another dataset, is a common risk, especially when variable number is large. The cross-validation strategy has been proven effective for the avoidance of overfitting (22,23). In this study, enrolled patients were randomly and equally split into five datasets for 5-fold cross-validation. For each running action, one of datasets was used as the testing group and the remaining four as the training group. This process repeated 5 times for each algorithm to find the optimal models. The performance of ML-based models was evaluated by the area under the receiver operating characteristic (ROC) curve (AUC) for predictive ability and the decision curve for clinical usefulness.
Feature selection
A classifier-specific evaluator for feature contribution was applied to each model to select variables. The potential models with best predictive performance and clinical usefulness were picked up to identify predictive risk factors. A list of variables sequenced by predictive contribution to the models was returned. Lower rank indicated better relevance to the model.
Statistical analysis
Univariate analysis was performed using SPSS 25.0 (IBM, New York, USA). Normality for quantitative data was analyzed by Shapiro-Wilk test. Normal quantitative parameters were compared under Student’s t-test and written as mean ± standard deviation (SD), while non-normal quantitative parameters were compared under Mann-Whitney U test and written as median with interquartile (IQR). Pearson’s Chi square test (or Fisher’s exact test when necessary) was used to compare the distribution of categorical variables. ML-based models were developed using Python programming language (version 3.7). Decision curve analysis (DCA) was performed using R software (version 3.6.3). Statistical significance was considered as P value <0.05 (two-side).
Results
Patient characteristics
Table 1 lists the clinical characteristics of all 709 patients involved in this study. The patients aged from 51 to 64 with a median age of 58 years old. LNM was observed in 80 (11.3%) patients. The node-positive group had a median CEA concentration of 3.63 ng/ml, significantly higher than node-negative group, indicating that a higher serum CEA level could be a risk factor of LNM. Additionally, a larger tumor size was significantly with LNM (P<0.001). In terms of the radiologic characteristic of lung cancer foci, node-positive group and node-negative group were significantly different in tumor density (P<0.001) and pleural indentation (P=0.02), but not in spiculation (P=0.315), vessel convergence (P=0.226) or lobulation (P=0.154). There was no pGGO cancer lesion in node-positive group. Further, analysis of clinicopathological features showed that the presence of micropapillary component (P<0.001), solid component (P<0.001), acinar component (P=0.001), LVI (P<0.001) and VPI (P<0.001) could be possible risk factors of LNM, while the presence of lepidic component indicated LNM-free disease (P<0.001). All node-positive patients were proved to be invasive adenocarcinomas by pathology.
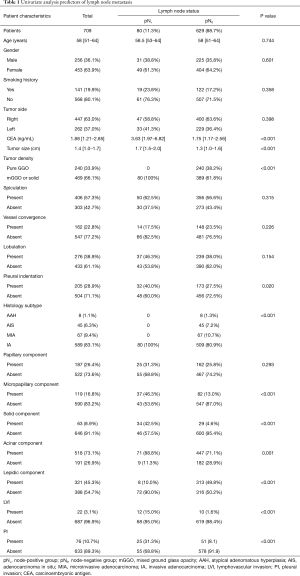
Full table
Predictive performance of ML-based models
Six supervised ML algorithms were used to develop efficient and reliable predictive models with 19 clinicopathological variables, and their predictive performance is illustrated in Figure 1 and Table 2. Among them, RFC model gave the best predictive performance (AUC =0.921, SD =0.014), closely followed by GBDT (AUC =0.919, SD =0.014), XGBoost (AUC =0.917, SD =0.028) and ANN (AUC =0.915, SD =0.017). As for two conventional methods, LR also performed well (AUC =0.935, SD =0.013), while the performance of MNB (AUC =0.876, SD =0.023) was poor. The sensitivity and specificity of different predictive models are given in Table S1.
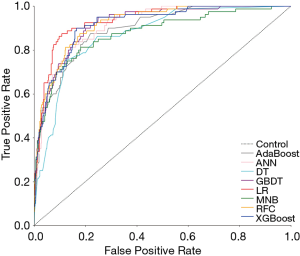
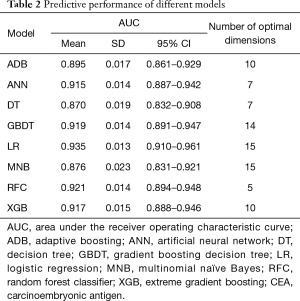
Full table
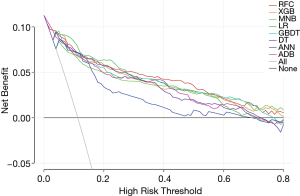
To further compare the clinical usefulness of models, DCA was performed (Figure 2). Firstly, across almost the entire reasonable range of thresholds, all models performed better than the two extreme lines (treat-all and treat-none lines). Most of them showed similar net benefits under most circumstances except for DT model. At the thresholds <0.28, LR presented slightly higher net benefits than other models. However, when the thresholds ≥0.28, RFC model performed best at most values of threshold probability. At the threshold range of 0–0.4, MNB performed almost worst among all models except DT. When thresholds >0.4, the net benefits of ADB and ANN decreased sharply and were lower than other models except DT. Therefore, in addition to RFC and LR, XGB and GBDT, which showed stably higher net benefits than other four models, were also identified as potential models.
Variable importance
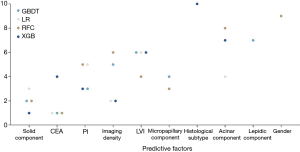
Based on four potential models (RFC, LR, XGB and GBDT) with great predictive performance and clinical usefulness, the top 10 important variables for LNM prediction and their rank are shown in Figure 3. The solid component ranked top to be the most influential predictive factor, followed by CEA, pleural invasion, tumor imaging density, LVI, micropapillary component, histological type, acinar component, lepidic component and gender, respectively.
Development of a dynamic predictive application
RFC model was considered the optimal model because of its excellent performance in both ROC curve and decision curve, which reached a high AUC with the minimal number of variables introduced, including CEA, solid component, micropapillary component, LVI and PI. Thus, a dynamic application of RFC model with these 5 variables was developed for the convenience of clinicians and patients (24).
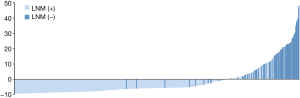
According to the application, the optimal cutoff point of risk probability to distinguish LNM (+) from LNM (−) was 13.85% (sensitivity: 87.5%; specificity: 82.2%). Figure 4shows the risk probability distribution of all patients, which has been standardized by the following formula: (risk probability−13.85%)/standard deviation.
Discussion
LNM status is crucial for the treatment of early-stage NSCLC. To date, lobectomy plus systematic LND is the standard management to achieve low recurrence rate and prolong survival (3,25). However, compared with selective LND or lymph node sampling, systematic LND could be more likely to cause a series of postoperative complications (26,27). On other occasions, sublobar resection including segmentectomy and wedge resection has been recommended for early-stage NSCLC patients, which showed similar survival outcome as lobectomy (28,29) and could also preserve more lung function. However, the sublevel surgery as selective LND and sublobar resection could more possibly lead to tumor residual and thus a poor prognosis if LNM occurred. Moreover, occult LNM makes the situation more complicated. It has been estimated that the occurrence rate of OLNM could be between 10.8% to 17.2% among stage I lung cancer (6-8). Patients with LNM might mistakenly undergo sublevel surgery, leading to a poor prognosis. For these patients, salvage management might be necessary. Therefore, more efforts should be given to accurately predict the LNM status during or after the operation.
Previous studies have revealed some possible predictive factors for LNM in NSCLC. Yu et al. reported several independent risk factors including tumor size, pleural invasion, and CEA (5). Pani et al. found that histologic subtypes could be related to lymph node status (4). Another similar study suggested different LND strategy for different combination of various clinicopathological features and CEA concentration and albumin level (30). These studies used uni- and multivariate analysis to reveal clinicopathological predictors for different LNM patterns. Our study, however, innovatively adopts ML algorithms to predict LNM by incorporating a large series of clinicopathological features. Among the predictive models, we found that RFC, GBDT, XGB, ANN all achieved AUC higher than 0.9, which was similar with LR model. However, in the decision curve, LR performed better than others at threshold <0.28, while RFC performed the best at most points of thresholds ≥0.28 and always kept a stably high net benefit. It is noteworthy that all models performed significantly better than treat-all and treat-none lines, indicating our models had clinical practice values and patients could gain more benefits if corresponding managements were conducted according to the predictive outcome of these models.
Furthermore, based on four potential models we identified with great performance in both ROC and decision curves, the top ten variables were found out, including solid component, CEA, pleural invasion, tumor imaging density, LVI, micropapillary component, histological subtype, acinar component, lepidic component and gender. In addition to CEA and imaging density that have been reported by previous studies (4,5), many histological features were also strongly related to the occurrence of LNM. Besides pleural invasion and LVI, histological details of growth such as the presence of solid, micropapillary and acinar components indicated high risk for LNM, while the presence of lepidic component could indicate LNM-free disease. In fact, these variables are conventionally not included in intraoperative pathology report. Our study emphasizes the importance of these histological features in the prediction of lymph node status. Thus, intraoperative pathology may be considered to include more detailed information about adenocarcinomas to further evaluate LNM risk, especially for patients who are hard to decide between lobectomy and sublobar resection. Importantly, the risk evaluation of LNM after surgery might be necessary for early-stage adenocarcinoma patients. For those who received sublobar resection or sublevel LND, the salvage management and close follow-up could be required if a high risk for LNM was observed based on our ML model.
In recent years, predicting metastasis with ML algorithms, as a promising alternative for other invasive or noninvasive diagnostic method, has been proven to be feasible in lung adenocarcinoma and colorectal cancer (11,12). These studies predicted on CT image and histologic evidence and obtained satisfying results. However, considered the sample size in the two study was not large, the validity of ML prediction needs to be further confirmed on a larger NSCLC patient population. Another methodological problem remained to be further explained is that the false-positive and false-negative rate need to be low enough to achieve good clinical utility. High AUC in ROC represents high predictive accuracy but does not necessary prove good clinical utility, because false-positive or false-negative results could reduce net benefit (31). To seek for a model that has high predictive accuracy and net benefit, we adopted DCA which has been widely proven to be efficiently and interpretable in the evaluation of clinical utility (32). From the decision curve, it was clear that RFC has the highest net benefit across the longest stable range of clinically reasonable preferences.
To further enhance the clinical usefulness of our study, a dynamic application of RFC model with 5 clinicopathological variables introduced was developed. So, clinicians and patients worldwide can benefit from our study and evaluate the LNM risk easily. The node-positive patients could be precisely identified by the RFC application (sensitivity: 87.5%; specificity: 82.2%; Figure 4).
This study is not without limitation. First, the ML algorithms can only yield dichotomous results (positive LNM or negative LNM), while hey cannot separate N1, N2, or N3. Second, the nature of retrospective analysis inevitably causes data acquisition bias. Lastly, the enrolled patients are from a single center and share an ethnicity. Future study is expected to validate the predictive performance of RFC model and more possible clinicopathological variables in a multicenter population.
Conclusions
This study comprehensively evaluated various ML-based predictive models and identified RFC model as the optimal one that accurately predicted LNM in early-stage adenocarcinomas. By feature selection, some clinicopathological characteristics were found to be strongly related to LNM. The pGGO or non-invasive adenocarcinoma (AAH, AIS and MIA) cancer lesion might indicate LNM-free disease, which also consisted with the presence of lepidic component. The application of RFC model was developed with great predictive ability and clinical usefulness. Thus, it can be feasible to evaluate the risk of LNM in patients with early-stage adenocarcinoma during or after the operation for clinical decision-making.
Acknowledgments
Funding: This work was supported by CAMS Innovation Fund for Medical Sciences (2017-12M-1-009; 2019-I2M-1-001).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-98
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-98
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-98). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Peking Union Medical College Hospital (No. S-K1049) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Lonardo F, Li X, Kaplun A, et al. The natural tumor suppressor protein maspin and potential application in non small cell lung cancer. Curr Pharm Des 2010;16:1877-81. [Crossref] [PubMed]
- De Leyn P, Dooms C, Kuzdzal J, et al. Revised ESTS guidelines for preoperative mediastinal lymph node staging for non-small-cell lung cancer. Eur J Cardiothorac Surg 2014;45:787-98. [Crossref] [PubMed]
- Pani E, Kennedy G, Zheng X, et al. Factors associated with nodal metastasis in 2-centimeter or less non-small cell lung cancer. J Thorac Cardiovasc Surg 2020;159:1088-96.e1. [Crossref] [PubMed]
- Yu X, Li Y, Shi C, et al. Risk factors of lymph node metastasis in patients with non-small cell lung cancer </= 2 cm in size: A monocentric population-based analysis. Thorac Cancer 2018;9:3-9. [Crossref] [PubMed]
- Kaseda K, Asakura K, Kazama A, et al. Risk Factors for Predicting Occult Lymph Node Metastasis in Patients with Clinical Stage I Non-small Cell Lung Cancer Staged by Integrated Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography. World J Surg 2016;40:2976-83. [Crossref] [PubMed]
- Park SY, Yoon JK, Park KJ, et al. Prediction of occult lymph node metastasis using volume-based PET parameters in small-sized peripheral non-small cell lung cancer. Cancer Imaging 2015;15:21. [Crossref] [PubMed]
- Moon Y, Choi SY, Park JK, et al. Risk Factors for Occult Lymph Node Metastasis in Peripheral Non-Small Cell Lung Cancer with Invasive Component Size 3 cm or Less. World J Surg 2020;44:1658-65. [Crossref] [PubMed]
- Pavlou M, Ambler G, Seaman S, et al. Review and evaluation of penalised regression methods for risk prediction in low-dimensional data with few events. Stat Med 2016;35:1159-77. [Crossref] [PubMed]
- Waljee AK, Higgins PD. Machine learning in medicine: a primer for physicians. Am J Gastroenterol 2010;105:1224-6. [Crossref] [PubMed]
- Zhong Y, Yuan M, Zhang T, et al. Radiomics Approach to Prediction of Occult Mediastinal Lymph Node Metastasis of Lung Adenocarcinoma. AJR Am J Roentgenol 2018;211:109-13. [Crossref] [PubMed]
- Takamatsu M, Yamamoto N, Kawachi H, et al. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput Methods Programs Biomed 2019;178:155-61. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- . Shalabi. LA, Shaaban. Z, Kasasbeh. B. Data Mining: A Preprocessing Engine. J Comput Sci 2006;2:735-9. [Crossref]
- Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol 2019;20:e262-73. [Crossref] [PubMed]
- Gonzalez GH, Tahsin T, Goodale BC, et al. Recent Advances and Emerging Applications in Text and Data Mining for Biomedical Discovery. Briefings in Bioinformatics 2016;17:33-42. [Crossref] [PubMed]
- Breiman L. Random Forests. Machine Learning 2001;
- Freund Y, Mason L. The Alternating Decision Tree Learning Algorithm. Morgan Kaufmann 2002.
- Freund Y, Schapire R. editors. A Short Introduction to Boosting 1999.
- Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. the 22nd ACM SIGKDD International Conference 2016.
- Cook JA, Ranstam J. Overfitting. Br J Surg 2016;103:1814. [Crossref] [PubMed]
- Jung Y. Multiple predicting K-fold cross-validation for model selection. Journal of Nonparametric Statistics 2018;30:197-215. [Crossref]
- Wu Y, Chong Y, Liu J, et al. Calculation tool for predicting the risk of lymph node metastasis in lung adenocarcinoma. Available online: https://nmgrmshinyappszzypumch.shinyapps.io/Pathology/. 2020. Accessed May 15 2020.
- Ginsberg RJ, Rubinstein LV. Randomized trial of lobectomy versus limited resection for T1 N0 non-small cell lung cancer. Lung Cancer Study Group. Ann Thorac Surg 1995;60:615-22; discussion 622-3. [Crossref] [PubMed]
- Han H, Zhao Y, Chen H. Selective versus systematic lymph node dissection (other than sampling) for clinical N2-negative non-small cell lung cancer: a meta-analysis of observational studies. J Thorac Dis 2018;10:3428-35. [Crossref] [PubMed]
- Okada M, Sakamoto T, Yuki T, et al. Selective mediastinal lymphadenectomy for clinico-surgical stage I non-small cell lung cancer. Ann Thorac Surg 2006;81:1028-32. [Crossref] [PubMed]
- Cao J, Yuan P, Wang Y, et al. Survival Rates After Lobectomy, Segmentectomy, and Wedge Resection for Non-Small Cell Lung Cancer. Ann Thorac Surg 2018;105:1483-91. [Crossref] [PubMed]
- Altorki NK, Yip R, Hanaoka T, et al. Sublobar resection is equivalent to lobectomy for clinical stage 1A lung cancer in solid nodules. J Thorac Cardiovasc Surg 2014;147:754-62; discussion 762-4. [Crossref] [PubMed]
- Zhao F, Zhen FX, Zhou Y, et al. Clinicopathologic predictors of metastasis of different regional lymph nodes in patients intraoperatively diagnosed with stage-I non-small cell lung cancer. BMC Cancer 2019;19:444. [Crossref] [PubMed]
- Zhang Z, Rousson V, Lee WC, et al. Decision curve analysis: a technical note. Ann Transl Med 2018;6:308. [Crossref] [PubMed]
- Vickers AJ, van Calster B, Steyerberg EW. A simple, step-by-step guide to interpreting decision curve analysis. Diagn Progn Res 2019;3:18. [Crossref] [PubMed]


