
Developing of risk models for small solid and subsolid pulmonary nodules based on clinical and quantitative radiomics features
Introduction
The International Agency for Research on Cancer reported that lung cancer was the most commonly diagnosed cancer (11.6% of the total cases) and the leading cause of cancer death (18.4% of the total cancer deaths) for both sexes in 2018 (1). Hence, efficient strategies are necessary to manage the disease. Early stage lung cancer usually presents as a solitary pulmonary nodule on thoracic computed tomography (CT), namely, a rounded lesion measuring less than 30 mm in diameter, which is completely surrounded by pulmonary parenchyma without other pulmonary abnormalities (2). According to its texture, nodules can be classified into subsolid nodules (SSNs) or solid nodules (3). Accurate characterization of pulmonary nodules, especially regarding their likelihood of malignancy, can be very important in lung cancer management.
Traditionally, thoracic radiologists and clinicians rely largely on qualitative morphological features such as texture, spiculation, lobulation, calcification, pattern of enhancement, presence of blood vessels, impact on adjacent structures, and so on, to evaluate the nature of pulmonary nodules (4). For example, a smoothly marginated solid nodule with internal fat and calcification can be a hamartoma, and a solid triangular subpleural nodule with a linear extension to the pleural surface is typical of an intrapulmonary lymph node, where no further CT follow-up is recommended (3). In addition, nodule size, nodule type, spiculation, upper lobe location, 18F-fluorodeoxyglucose uptake, and volume doubling time (VDT) could be independent predictors of malignancy (5-8). However, morphological features can sometimes overlap between benign and malignant nodules, so the morphological features are not sufficient to assess the likelihood of malignancy (9). In addition, inter- and intraobserver agreement in evaluating nodule features was found to be highly variable (10). Moreover, nodules less than 10 mm in size, which account for approximately 80% of all non-calcified nodules, are less amenable than larger nodules when it comes to characterization (11).
At present, a rapidly evolving field called radiomics, which enables digital decoding of images into quantitative features, including descriptors of size, shape, and textural features, has shown promise in characterizing lung cancers (12). For example, previous radiomics studies were successfully performed to identify epidermal growth factor receptor mutations, classify tumor histologic subtype, and predict tumor invasiveness as well as lymph node and distant metastasis (13). Therefore, the current study aimed to develop risk models for small solid and subsolid pulmonary nodules (5–20 mm) based on clinical and quantitative radiomics features.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-80).
Methods
Patient enrollment
Data were retrospectively collected from the West China Hospital of Sichuan University between January 2010 and July 2017. Patients were enrolled if there was an untreated, pathologically confirmed, 5–20 mm, dominant, noncalcified SSN, or solid nodule on thoracic high-resolution CT (HRCT). The study included incidental pulmonary nodules and annually screen detected nodules. Patients were excluded if (I) multiple pulmonary nodules were observed, where nodules distributed diffusely or multiple nodules were confirmed to be tumors or if there were several problematic nodules; (II) pleural effusion, atelectasis, or lymph node enlargement was observed; and (III) the pathological diagnosis was not clear or it was a metastatic tumor. Pathological diagnoses of all nodules were based on sputum cytology or pathologic examination of lung tissues obtained from bronchoscopy, CT-guided percutaneous lung biopsy, or thoracic surgery. In total, 1,855 patients with 5–20 mm pathologically confirmed nodules were found, and 1,171 patients were enrolled for further analysis, as the segmentation of two patients failed and 682 patients did not have HRCT.
Collection of clinical variables
Clinical characteristics were recorded, including age, sex, smoking status, history of malignancy, and family history of lung cancer (demographic); diameter, location, shape, spiculation, lobulation (radiologic), red blood cells, white blood cells, blood platelets, neutrophil to lymphocyte ratio (NLR), prothrombin time, activated partial thromboplastin time (APTT), carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CYFRA21-1) and neuron specific enolase (NSE, laboratory). This study was approved by the institutional review board of the West China Hospital of Sichuan University, and the requirement to obtain informed consent was waived as the privacy and identity information of the subjects were guaranteed. There were some missing data regarding CEA, CYFRA21-1, and NSE tests, which were populated by the median value. The data were not available as some of the patients did not undergo testing.
CT image acquisition
The CT image chosen was the one prior to the initial treatment. All images were acquired using several models of multi-row spiral CT scans from GE, Siemens or Philips scanners. Most CT scans were obtained between 100 and 120 kV. The pixel size was 0.7 to 0.9 mm, and the axial slice thickness was less than 1.25 mm, with 1 mm being the most common. Reconstruction was performed using a standard convolution kernel. Some patients underwent non-contrast CT examination while others underwent contrast-enhanced CT. All images were analyzed in both lung (width, 1,500 HU; level, −700 HU) and mediastinal (width =350 HU; level =40 HU) settings. During patient selection and radiologic feature evaluation, two specialists who were blinded to the pathological results of lesions evaluated all CT scans and resolved discrepancies by consensus.
Nodule segmentation and feature extraction
The target nodules were manually segmented in 3D using the ITK-SNAP software by one clinician in pulmonary and critical care medicine, with four years of experience in pulmonary nodule evaluation (14). The author was also blinded to the pathological results of all the lesions. Radiomics feature extraction was performed using the open-source platform Pyradiomics (version 2.0.0), which enables quantitative features to be extracted from HRCT images (12). Extracted from Pyradiomics were 14 shape features, 18 first-order features, 22 gray-level co-occurrence matrix (GLCM) features, 16 gray-level size zone matrix (GLSZM) features, 16 gray-level run length matrix (GLRLM) features, and 14 gray-level dependence matrix (GLDM) features. Except for shape features, other features were also calculated from the filtered images (LoG with five sigma levels and wavelet with eight derived images). In total, 1,218 radiomics features [14 shape features + 86 other features × (1 original image + 13 filtered images)] were generated for each nodule. A detailed list of the extracted features is provided in Table S1.
Model establishment
All risk models were established in two steps: feature selection and model development. The least absolute shrinkage and selection operator (LASSO) method was applied to select the most significant predictors from clinical and radiomics features simultaneously. Then, four types of models were tried for each nodule group based on selected features, including random forest, XGBOOST, SVM, and logistic model. We normalized the data using the Z-score to allow all the coefficients to be based on the same scale and performed five-fold cross-validation during both steps.
Performance of models
The model performance was evaluated using metrics such as the area under the ROC curve (AUC), accuracy, F1 score, recall, precision, sensitivity, and specificity. The performance of the models was compared with the predictive ability of the Mayo model (5). All cases in each group were utilized to validate the Mayo model, and the corresponding AUC, accuracy, sensitivity, and specificity were calculated.
Statistical analysis
The numerical data were presented as mean ± standard deviation and compared using Student’s t-test, while categorical data were described as percentages and compared using the chi-square test. All statistical tests were two-sided, and differences were considered statistically significant at P<0.05. All statistical analyses were performed with R version 3.6, and all models were established using Python 3.7.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of the West China Hospital of Sichuan University (No.59). Informed consent was waived as this was a retrospective study and the privacy and identity information of the subjects were guaranteed.
Results
Patient enrollment and pathology of nodules
Figure 1 presents the flowchart of the study process. A total of 1,171 patients who met the inclusion criteria were enrolled, including 548 patients with solid nodules (malignant, 242; benign, 306) and 623 patients with SSNs (malignant, 519; benign, 104). All nodules were confirmed pathologically. The benign group included inflammatory nodules (62.20%), benign tumors (20.00%), and other lesions (17.80%). Malignant nodules were mainly adenocarcinomas (97.37%), while 1.97% were squamous carcinomas and 0.66% were other types (Table S2).
Patient clinical characteristics
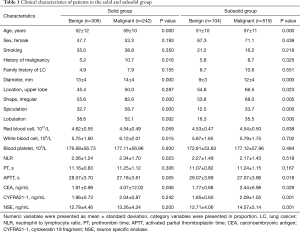
The clinical characteristics of the patients are summarized in Table 1. In patients with solid nodules, the benign and malignant groups exhibited some differences in age, history of malignancy, diameter, shape, spiculation, lobulation, white blood cell count, NLR, APTT, and CEA (P<0.05). As for SSNs, there was no difference in the proportion of history of malignancy, white blood cell count, NLR, and CEA between the benign and malignant groups. However, significant differences in age, diameter, location, shape, spiculation, lobulation, APTT, CYFRA21-1, and NSE were observed between the malignant and benign groups (P<0.05). Details of missing data for CEA, CYFRA21-1, and NSE are described in Table S3.
Full table
Model establishment and selected radiomics features
We developed lung cancer risk models among four nodule groups: all nodules (n=1,171), nodules ≤10 mm (n=409), solid nodules (n=548), and SSNs (n=208). To avoid overfitting of models in the SSN group, only 104 malignant SSNs were included, which were 1:1 matched with benign SSNs.
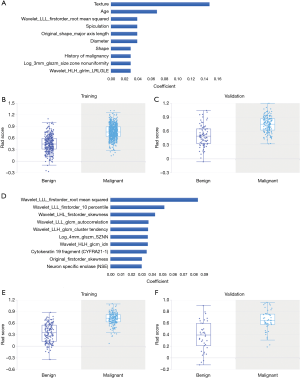
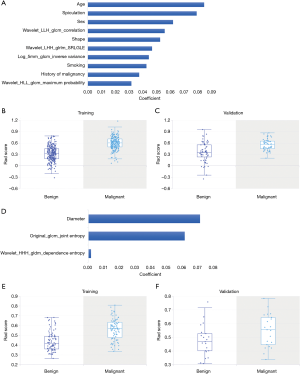
Table S4 summarizes the selected features from LASSO analysis in detail for all nodules (α=0.00955, n=74 features), nodules ≤10 mm (α=0.0152, n=67 features), solid nodules (α=0.0152, n=51 features), and SSNs (α=0.12328, n=3 features). Figure 2 describes the top ten features sorted by the absolute value of coefficients and the rad-score distribution based on selected features for all nodules (Figure 2A,B,C) and nodules ≤10 mm (Figure 2D,E,F). In addition, the top ten features for solid nodules, all three features for SSNs, and the corresponding rad-score distribution for solid nodules (Figure 3A,B,C) and SSNs (Figure 3D,E,F) are shown in Figure 3. There was a higher proportion of radiomics features selected for nodules ≤10 mm (83.6% of 67 features and 80% of the top ten features).
Performance of risk models
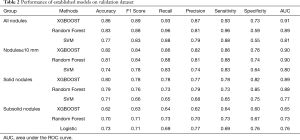
We performed five-fold cross-validation to evaluate the predictive performance of the lung cancer risk models. According to the accuracy of the training dataset, the top three models for each nodule group were recorded. For all nodules (410 benign vs. 761 malignant; 1:1.86), nodules ≤10 mm (185 benign vs. 224 malignant; 1:1.21), and solid nodules (306 benign vs. 242 malignant; 1.26:1), the top three models were XGBOOST, random forest, and SVM, respectively. However, it was the XGBOOST, random forest, and logistic model for SSNs (104 benign vs. 104 malignant; 1:1). The performance of the established models on the validation dataset is summarized in Table 2, while that of the training dataset is shown in Table S5.
Full table
In addition, the ROC curves of the top three models in the training and validation datasets, as well as the performance of the best model in each fold on the validation dataset are shown in Figure 4 (all nodules, Figure 4A,B,C; nodules ≤10 mm, Figure 4D,E,F) and Figure 5 (solid nodules, Figure 5A,B,C; SSNs, Figure 5D,E,F). The performance of risk models for SSNs was quite weak (AUC in validation dataset, 0.65–0.76) and the XGBOOST model was even unstable (AUC, 1.00 in training vs. 0.65 in validation); therefore, the ROC curves of the random forest model are shown in Figure 5F.
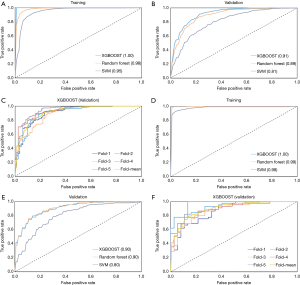
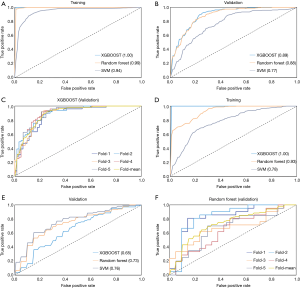
Comparison of established clinical-radiomics models with Mayo model
The Mayo model was validated in all cases in each nodule group and was compared with the best corresponding clinical-radiomics model. Table 3 summarizes the detailed predictive results. We found that the Mayo model, which was based on only six clinical characteristics, showed comparable discriminating ability with our clinical-radiomics model for SSNs (accuracy, 0.73 vs. 0.70; AUC, 0.75 vs. 0.73). However, it was inferior to the clinical-radiomics models for the other three nodule groups (accuracy, 0.55–0.67 vs. 0.80–0.86; AUC, 0.59–0.70 vs. 0.89–0.91).

Full table
Discussion
Accurate evaluation of the malignancy of pulmonary nodules plays an important role in lung cancer management. Moreover, solid nodules and SSNs exhibit different clinical courses, and guidelines for the management of pulmonary nodules have provided separate recommendations for the two types of nodules (9). Therefore, the current study based on clinical and radiomics features has established four models for all nodules (5–20 mm), nodules ≤10 mm, solid nodules, and SSNs.
We previously established lung cancer risk models solely based on the clinical features of solid nodules and SSNs of different sizes (15). The clinical models showed an AUC of 0.70 and 0.71 for SSNs and solid nodules <15 mm, 0.72 and 0.81 for SSNs and solid nodules between 15 and 30 mm, respectively (15). It was obvious that the current models combining clinical and radiomics features exhibited greater discrimination ability, especially for nodules ≤10 mm (highest AUC, 0.90) and solid nodules (highest AUC, 0.89), while there was only a slight improvement for SSNs (highest AUC, 0.76). Similarly, when compared to the Mayo model (six predictors: age, smoking, history of cancer, diameter, spiculation, and upper lobe location), the established models demonstrated absolute superiority for all nodules, nodules ≤10 mm, and solid nodules, whereas the diagnostic advantages for SSNs were not obvious. The possible reason was that there were only a small number of benign SSNs available, and for data balance, the same number of malignant cases were matched. Therefore, due to the small sample size (208), trained SSN models can be unstable, of which only three risk predictors were selected.
An increasing number of SSNs have been encountered in routine clinical practice. Most early stage lung adenocarcinomas can manifest as SSNs with different degrees of invasion (16). Consequently, previous radiomics studies for SSNs focused more on identifying the invasiveness of lung adenocarcinomas (16-18). However, Gong et al. studied 182 histopathology-confirmed SSNs using radiomics analysis for nodule diagnosis [59 benign nodules, 50 adenocarcinoma in situ (AIS), 32 minimally invasive adenocarcinoma (MIA), and 41 invasive adenocarcinoma] (19). Their results showed an average AUC of 0.75 in distinguishing benign and malignant SSNs, which was consistent with the current study (AUC, 0.65–0.76). Moreover, their models also demonstrated poor performance in benign and AIS nodules (AUC, 0.55), and benign and MIA nodules (0.77), respectively (19). Nevertheless, a high AUC of 0.93, was observed for benign nodules and invasive adenocarcinomas (19). Hence, radiomics features have shown potential in predicting the malignancy of SSNs, but sufficient data is a priority to train a good model.
We noticed that the clinical characteristics could be as important as radiomics features in lung cancer risk prediction for solid nodules. Sixty percent of the top ten selected features were clinical variables, which have been identified in previous studies (7,20-22). The clinical variables were age, spiculation, sex, shape, smoking, and history of malignancy. When predicting malignancy of solid nodules, the clinical-based models exhibited an AUC of 0.81 to 0.89 (7,20-22), and one study pointed out that a VDT of 25–400 days was highly suggestive of malignancy (7). On the other hand, quantitative radiomics models have also demonstrated potential for diagnosing solid nodules, especially radiomics models created from gross tumor volume instead of peritumoral volumes (23). Therefore, the combination of clinical and quantitative radiomics features improved the performance of the clinical models developed in our previous study (15).
The Brock model, which is based on two screening cohorts (PanCan and BCCA; rates of cancer, 5.5% and 3.7%, respectively) demonstrated good discrimination and calibration for nodules ≤10 mm, with an AUC of 0.89 to 0.94 (8). Predictors of cancer in the model were all clinical variables, including older age, female sex, family history of lung cancer, emphysema, larger nodule size, upper lobe location, part-solid nodule type, lower nodule count, and spiculation (8). Regarding incidental pulmonary nodules ≤10 mm, Xu et al. predicted the malignancy of 127 sub-centimeter nodules using radiomics features and achieved an AUC of 0.84 (24). However, in the current study of incidental pulmonary nodules, 67 clinical and radiomics features were selected for nodules ≤10 mm, and the risk models showed the highest AUC of 0.90. It is worth noting that radiomics features played a more important role in lung cancer risk prediction for incidental nodules ≤10 mm, with a high proportion of 80% among the top ten features, which were completely different from the other three nodule groups. Hence, radiomics features are meaningful in malignancy prediction for nodules ≤10 mm in routine clinical practice.
This study had some general and study-specific limitations. First, the study was carried out in a single medical center, in which bias could exist. In addition, this was a retrospective case-control study, which resulted in different scanner machines and non-uniform imaging protocols across patients. The difference should have a relevant impact on radiomics feature stability and thus prevented the establishment of robust risk models. Second, the growth rate of pulmonary nodules is one of the key characteristics associated with lung cancer probability, but we failed to apply the relevant parameters to build risk models in the current study. Usually, at least two thoracic CT scans prior to treatment are needed to calculate the growth rate of a nodule, but most of the patients we studied did not meet the criteria. Third, only a small number of SSNs were available for modeling; therefore, the performance of the established SSN models was unstable. Increasing the sample size, especially for benign SSNs, is warranted in future research. Last but not the least, we performed five-fold cross validation to evaluate the performance of established risk models, whereas the external validation is necessary.
Conclusions
In conclusion, based on both clinical and radiomics features, the current study established risk models to predict the malignancy of 5–20 mm pulmonary nodules. The models were developed for four nodule groups, including all nodules, nodules ≤10 mm, solid nodules, and SSNs. All models demonstrated excellent discrimination ability except for those of SSNs. Further studies are warranted to develop robust SSN models.
Acknowledgments
Funding: This work was supported by the National Key Development Plan for Precision Medicine Research (2017YFC0910004), and Central Guide Place-Free Exploration Project, Sichuan Provincial Department of Science and Technology (2020ZYD005).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-80
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-80
Peer Review File: Available at https://dx.doi.org/10.21037/jtd-21-80
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-80). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of the West China Hospital of Sichuan University (No.59). Informed consent was waived as this was a retrospective study and the privacy and identity information of the subjects were guaranteed.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Bai C, Choi CM, Chu CM, et al. Evaluation of Pulmonary Nodules: Clinical Practice Consensus Guidelines for Asia. Chest 2016;150:877-93. [Crossref] [PubMed]
- MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. [Crossref] [PubMed]
- Wu W, Pierce LA, Zhang Y, et al. Comparison of prediction models with radiological semantic features and radiomics in lung cancer diagnosis of the pulmonary nodules: a case-control study. Eur Radiol 2019;29:6100-8. [Crossref] [PubMed]
- Swensen SJ, Silverstein MD, Ilstrup DM, et al. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med 1997;157:849-55. [Crossref] [PubMed]
- Herder GJ, van Tinteren H, Golding RP, et al. Clinical prediction model to characterize pulmonary nodules: validation and added value of 18F-fluorodeoxyglucose positron emission tomography. Chest 2005;128:2490-6. [Crossref] [PubMed]
- Soardi GA, Perandini S, Motton M, et al. Assessing probability of malignancy in solid solitary pulmonary nodules with a new Bayesian calculator: improving diagnostic accuracy by means of expanded and updated features. Eur Radiol 2015;25:155-62. [Crossref] [PubMed]
- McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369:910-9. [Crossref] [PubMed]
- Kim TJ, Kim CH, Lee HY, et al. Management of incidental pulmonary nodules: current strategies and future perspectives. Expert Rev Respir Med 2020;14:173-94. [Crossref] [PubMed]
- van Riel SJ, Sanchez CI, Bankier AA, et al. Observer Variability for Classification of Pulmonary Nodules on Low-Dose CT Images and Its Effect on Nodule Management. Radiology 2015;277:863-71. [Crossref] [PubMed]
- van't Westeinde SC, de Koning HJ, Xu DM, et al. How to deal with incidentally detected pulmonary nodules less than 10mm in size on CT in a healthy person. Lung Cancer 2008;60:151-9. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Liao Z, Mohan R, Court L, et al. Radiomics of pulmonary nodules and lung cancer. Sci Rep 2017;6:86-91. [PubMed]
- Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006;31:1116-28. [Crossref] [PubMed]
- Zhang R, Tian PW, Chen BJ, et al. Predicting Lung Cancer Risk of Incidental Solid and Subsolid Pulmonary Nodules in Different Sizes. Cancer Manag Res 2020;12:8057-66. [Crossref] [PubMed]
- Fan L, Fang M, Li Z, et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol 2019;29:889-97. [Crossref] [PubMed]
- Wu G, Woodruff HC, Shen J, et al. Diagnosis of Invasive Lung Adenocarcinoma Based on Chest CT Radiomic Features of Part-Solid Pulmonary Nodules: A Multicenter Study (vol 297, 2020192431, 2020). Radiology 2020;297:E282 [Crossref] [PubMed]
- Zhao W, Xu Y, Yang Z, et al. Development and validation of a radiomics nomogram for identifying invasiveness of pulmonary adenocarcinomas appearing as subcentimeter ground-glass opacity nodules. Eur J Radiol 2019;112:161-8. [Crossref] [PubMed]
- Gong J, Liu J, Hao W, et al. Computer-aided diagnosis of ground-glass opacity pulmonary nodules using radiomic features analysis. Phys Med Biol 2019;64:135015 [Crossref] [PubMed]
- Chen XB, Yan RY, Zhao K, et al. Nomogram For The Prediction Of Malignancy In Small (8–20 mm) Indeterminate Solid Solitary Pulmonary Nodules In Chinese Populations. Cancer Manag Res 2019;11:9439-48. [Crossref] [PubMed]
- Guo HY, Lin JT, Huang HH, et al. Development and Validation of a 18F-FDG PET/CT-Based Clinical Prediction Model for Estimating Malignancy in Solid Pulmonary Nodules Based on a Population With High Prevalence of Malignancy. Clin Lung Cancer 2020;21:47-55. [Crossref] [PubMed]
- She Y, Zhao L, Dai C, et al. Development and validation of a nomogram to estimate the pretest probability of cancer in Chinese patients with solid solitary pulmonary nodules: A multi-institutional study. J Surg Oncol 2017;116:756-62. [Crossref] [PubMed]
- Wu S, Zhang N, Wu Z, et al. Can Peritumoral Radiomics Improve the Prediction of Malignancy of Solid Pulmonary Nodule Smaller Than 2 cm? Acad Radiol 2020; Epub ahead of print. [Crossref] [PubMed]
- Xu Y, Lu L. Application of Radiomics in Predicting the Malignancy of Pulmonary Nodules in Different Sizes. AJR Am J Roentgenol 2019;213:1213-20. [Crossref] [PubMed]



