
Upregulation of estrogen receptor beta protein but not mRNA predicts poor prognosis and may be associated with enhanced translation in non-small cell lung cancer: a systematic review and meta-analysis
Introduction
Estrogen receptor beta (ERβ, also known as ESR2) is one of the two classical subtypes of the estrogen receptor (1) and is widely expressed in the lung. Lung cancer cells exhibit a stronger ERβ protein expression than normal lung cells (2-4). Previous studies, including ours, have reported that ERβ in the nucleus acts as a transcription factor that exerts effects on lung carcinogenesis (5,6); and ERβ in the cytosol participates in the signaling pathways that promote non-small cell lung cancer (NSCLC) progression and metastasis (7,8), leading to a poor prognosis in NSCLC patients.
Despite unraveling the putative role of ERβ in lung cancer, the prognostic value of ERβ protein expression reported in different studies is not consistent (9,10). A meta-analysis in 2015 reported a favorable effect of ERβ on NSCLC survival (11). Another meta-analysis of 11 studies reported that ERβ is not an independent predictor of NSCLC survival (12). These two meta-analyses lacked evaluation by different subcellular localizations of ERβ, and neither of these studies considered the effect of ERβ in lung adenocarcinoma. Several additional original studies were published subsequently.
Studies focusing on the expression and prognostic value of ERβ mRNA have shown negative results. ERβ mRNA expression is not upregulated in lung cancer tissues (13). In 2015, a meta-analysis showed that ERβ mRNA is not a prognostic factor for NSCLC (14). Therefore, we suppose that the expression of ERβ protein and mRNA is not consistent in NSCLC, and that this inconsistency is reflected in their prognostic values. There might be an abnormal translation control that can upregulate the protein expression of ERβ in the presence of low mRNA expression level, which further affects the prognosis. As the most abundant base modification of RNA, N6-methyladinosine (m6A) drives translation initiation in human cells (15). METTL3 acts as a key enzyme in RNA m6A modification. In 2016, Lin et al. (16) reported that METTL3 promotes the translation of abundant mRNAs in human cancer cells, revealing the important role of translation control in cancer cells.
The expression of ERβ and its relationship with lung cancer survival have not been systematically evaluated to date. Therefore, in this study, we conducted a meta-analysis with specific subgroups and a large sample size. By combining abundant transcriptome data, we illustrated the prognostic value of ERβ protein and mRNA. Expression data from clinical samples and public databases were included to explore the difference between ERβ protein and mRNA expression. Furthermore, we verified this difference in paired tissue samples from NSCLC patients. Finally, we explored the possible mechanism of METTL3-driven translation regulation in cell lines. In summary, based on our studies of estrogen-mediated NSCLC progression (7,8,17-19), we provide insights into ERβ expression profiles, and the possibility of anti-estrogen therapy in NSCLC.
We present the following article in accordance with the PRISMA reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-658) (20).
Methods
Literature search
We comprehensively searched the PubMed, Web of Science, and Embase databases for articles published before October 6, 2020 (the search strategy is detailed in Table S1) that analyzed the prognostic value of ERβ protein expression in NSCLC patients. We identified studies using medical subject headings (MeSH) from PubMed to develop a controlled vocabulary where applicable. In addition to MeSH, we included relevant free-text entry terms.
Selection criteria and study selection
Original articles that achieved all the following criteria were included: (I) presented data, including different histological types, from primary NSCLC patients; (II) detected ERβ protein expression in primary tumor tissues using immunohistochemistry (IHC); (III) provided detailed clinical and pathological features of the study population; (IV) addressed the prognostic value of tumor ERβ expression using survival analysis.
Articles that achieved any of the following criteria were excluded: (I) estimated the prognostic value for subtypes of ERβ only; (II) considered disease-free survival, recurrence-free survival, or progression-free survival as an endpoint but not overall survival (OS).
After removing duplicate studies, two investigators (W Meng and H Xiao) initially screened the titles and abstracts of the identified records for full-text review. Disagreements were resolved by consensus.
Data extraction
Using a pre-specified data collection sheet, two investigators (W Meng and H Xiao) independently extracted data from the retrieved articles, and agreement was achieved in group discussions. The extracted data included the authors, year of publication, country or region, sample size, tumor stage, composition of different histology types, median/mean and range of age, follow-up duration, epidermal growth factor receptor (EGFR) mutation status, aromatase expression status, sample type, detection method of ERβ expression, ERβ subcellular localization, positive cut-off definition, number of positive cases, antibody type, and covariates adjustment. We selected OS as the endpoint for our meta-analysis because OS is widely used as a significant prognostic indicator. Hazard ratios (HRs) for the association between three subcellular localizations of ERβ [cytoplasmic ERβ, nuclear ERβ, and overall ERβ (cytoplasmic and nuclear ERβ)] and OS were extracted.
Quality assessment
We used the Newcastle-Ottawa Scale (NOS) (21) for cohort studies to assess the methodological quality of the included studies. It is a nine-point scoring system that considers participant selection, exposure measurement, ascertainment of outcomes, covariate adjustment, and adequacy of follow-up. A high-quality study was defined as a study with at least seven points. Two reviewers (W Meng and H Xiao) independently assessed the quality of the included articles, and disagreements were resolved through group discussions.
Statistical analysis of meta-analysis
HRs and the corresponding 95% confidence intervals (CIs) were considered the effect sizes in this study. For studies in which HRs and 95%CIs were not available, we used the method described by Parmar et al. (22) to derive estimates from the published Kaplan-Meier survival curves. The most adjusted study-specific HRs and 95%CIs were primarily pooled using a random-effects model and the inverse variance method. Heterogeneity between studies was assessed using the I2 index, and we considered I2<50% as low heterogeneity, I2 between 50% and 75% as medium heterogeneity, and I2>75% as high heterogeneity. If the heterogeneity was low (I2<50%), a fixed-effects model was used. Subgroup analyses were performed based on several variables, including histology type, geographical location and whether a multivariate or univariate analysis was performed. Sex-specific HRs were also pooled using data from the studies. For study groups with relatively high heterogeneity (I2>50%), we performed sensitivity analysis (in the random-effects model) using a leave-one-out strategy to investigate the sources of heterogeneity.
Publication bias was assessed using funnel plots and the Egger’s test. All statistical analyses for the meta-analysis were performed using STATA version 16.0 (Stata Corp, College Station, TX, USA); a 2-sided P value of <0.05 was considered statistically significant.
Survival analysis of gene expression in lung adenocarcinoma
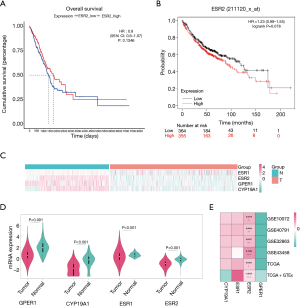
After the removal of 59 normal tissues, we analyzed the prognostic value of four genes [ESR2, ESR1 (estrogen receptor alpha; ERα), G-protein coupled estrogen receptor 1 (GPER1) and CYP19A1 (also known as aromatase, which is a key enzyme for estrogen synthesis)] in 526 lung adenocarcinoma tissues from The Cancer Genome Atlas (TCGA)-lung adenocarcinoma (LUAD). The “survminer” and “survival” packages were used to draw Kaplan-Meier curves. The Kaplan-Meier Plotter (http://kmplot.com/) is an online database used to assess the association of genes with survival in four types of cancer (lung, breast, gastric, and ovarian cancer). It was used to verify the prognostic value of the four target genes in lung adenocarcinoma patients (n=719).
Differential expression analysis of the transcriptome in lung adenocarcinoma
Based on the findings of the relationship between ERβ and lung cancer at the protein level using meta-analysis, we performed bioinformatics analysis to further explore the relationship between ERβ and lung cancer at the transcriptional level.
To quantify ERβ mRNA expression in lung adenocarcinoma, RNA-seq data of a lung adenocarcinoma cohort (tumor tissues =526 vs. normal tissues =59) from TCGA and those of normal lung tissues (n=288) from the Genotype-Tissue Expression (GTEx) were obtained from the University of California Santa Cruz Xena platform (https://xenabrowser.net/datapages/). RNA-seq data from the GTEx (https://commonfund.nih.gov/GTEx/) project were used to reduce the bias in the data from TCGA-LUAD. ESR2, ESR1, GPER1 and CYP19A1 were selected as the target genes for differential expression analyses. The R package “limma” was used to screen the differentially expressed genes (DEGs) among the four target genes. Genes with an absolute value of logFC (the logarithm of fold change) >1 and P<0.05 were defined as the DEGs. The results of “limma” analysis are presented in the form of a heatmap. Four gene expression profile datasets (GSE10072, GSE40791, GSE32863, and GSE43458) of lung adenocarcinoma were obtained from the Gene Expression Omnibus (GEO) for further validation of the above DEG analysis results.
Patients and tissue specimens
Paired samples of primary NSCLC tumors and corresponding normal adjacent tissues from 39 Chinese patients for IHC were obtained at the time of surgical resection at the Department of Thoracic Surgery, Tongji Hospital of Huazhong University of Science and Technology Tongji Medical College (Wuhan, China).
Paired samples of primary tumors and normal adjacent tissues from two patients for western blotting and RT-qPCR were obtained at the time of surgical resection at the Department of Thoracic Surgery, Union Hospital of Huazhong University of Science and Technology Tongji Medical College (Wuhan, China). The exterior of the NSCLC tissue which is a hard white part, was selected. The study was approved by the Ethics or Institutional Review Board of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, and written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki (as revised in 2013). All of the patients had sufficient tissue blocks available for the analysis of ERβ expression.
Immunohistochemical analyses
Sample processing and IHC were performed as previously described (17). Rabbit anti-human ERβ polyclonal antibody (dilution 1:50, Proteintech 14007-1-AP) was purchased from Proteintech (Wuhan, China). Protein expression levels were independently scored by two pathologists. Immunoreactivity scores of cancer tissue samples were determined based on staining intensity and positive staining area according to the method described by Tang et al. (17). Proportion of the positive cells was scored as follows: 1, ≤25% positive cells; 2, 25–50% positive cells; 3, 50–75% positive cells; and 4, >75% positive cells. Staining intensity was evaluated as follows: 1, negative; 2, weakly positive; 3, moderately positive; and 4, strongly positive. A score of 1–8 was obtained by adding the staining intensity score and the proportion score. A total score ≥5 was defined as high expression, and a score ≤4 was defined as low expression. These criteria were based on the evaluations reported by Nose et al. and Kawai et al. (23,24).
Cell lines and culture conditions
The human NSCLC cell line A549 was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), grown for 2 weeks, and passaged four times before freezing aliquots for subsequent analyses. The cell lines were tested and authenticated by ATCC. The cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and then incubated in a humidified atmosphere with 5% CO2 at 37 °C.
Cell transfection
Transfection of small interfering RNA (siRNA) was performed using Lipofectamine 3000 (Invitrogen; Shanghai, China) according to the manufacturer’s instructions. siRNAs targeting METTL3 (siRNA_METTL3: CTGCAAGTATGTTCACTATGA) and control siRNAs (siRNA_NC) were obtained from Ribobio Co., Ltd, Guangzhou China.
Western blotting of cultured cells and NSCLC samples
Lung cancer tissues or cell lines were lysed in RIPA buffer, followed by homogenization and determination of protein concentration. Thereafter, 20 µg of protein was loaded for SDS-PAGE and transferred onto PVDF membranes. Immunoblotting was performed to detect protein expression. The corresponding antibodies included METTL3 (Abcam 195352), ERβ (Proteintech 14007-1-AP), and GAPDH (Proteintech 60004-1-Ig). Specifically bound HRP-conjugated secondary antibodies were detected using an ECL detection system (ChemiDocTM XRS+ machine, Bio-Rad Laboratories). Densitometric analyses were performed using ImageJ software. Relative quantification was performed after normalization to GAPDH band intensities. A Mann-Whitney test was performed to assess the difference in protein expression between the groups. Each experiment was performed in triplicates and repeated at least three times.
RNA isolation and RT-qPCR
Total RNA was extracted from NSCLC tissues (preserved in RNA Keeper Tissue Stabilizer, Vazyme Biotech Co. LTD, Nanjing, China) and cells using TRIzol (Invitrogen), following the manufacturer’s instructions. 1 µg total RNA was reverse transcribed using SuperScript® III Reverse Transcriptase (Invitrogen). Quantitative RT-PCR was performed using SYBR® Green PCR Master Mix with the StepOne Real-Time PCR System (Applied Biosystems). GAPDH was used as the internal control for the normalization. The primers were purchased from Tsingke Co. LTD, Beijing, China. Primer information from PrimerBank is shown in Table S2.
Results
Literature search
Electronic searches identified 2,845 citations from PubMed, Web of Science and Embase, of which 1,764 titles and abstracts were reviewed. After the excluding 1,688 records, the full texts of 76 articles were further reviewed (Figure 1). A total of 23 unique studies in 22 articles met our selection criteria and were included in the meta-analysis.
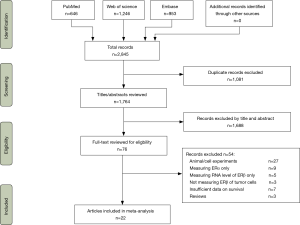
Characteristics and quality of the included studies
The characteristics of the included studies are summarized in Table 1. The included studies reported data from a total of 3,744 patients. All studies were published between 2005 and 2020, including 22 retrospective cohort studies and one prospective cohort study. Fifteen studies from fourteen articles were on NSCLC, and eight studies were on lung adenocarcinoma (25-32).
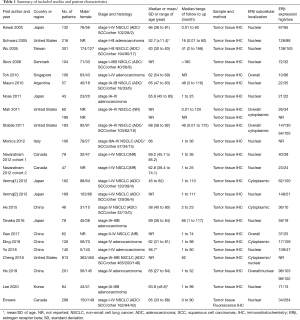
Full table
All studies measured the protein expression of ERβ in tumor tissue using IHC. Six studies included cytoplasmic ERβ (2,10,25,33-35), 16 studies included nuclear ERβ (9,10,23,26-29,31,32,36-40), and five studies included overall ERβ, which considered cytoplasmic and nuclear ERβ expression (2,30,32,34,41). Four studies reported two HRs for these different subcellular localizations of ERβ (2,10,32,34). Navaratnam et al. included two distinct studies that reported two HRs for nuclear ERβ (38). Eight studies performed multivariate analyses of HRs to adjust for sex, age, stage at diagnosis, or smoking status (2,9,10,28,34,39,42). Fourteen studies were performed in East Asia (9,23,25-27,29-33,35,40,41), six in North America (2,10,28,34,38,42), two in Western Europe (37,39) and one in South America (36). The tumor stages of patients in most studies were stage I–IV or I–III. He (in 2015) et al. (33) and He (in 2019) et al. (32) studied stage IV lung cancer patients only. Monica et al. (37) studied stage III-IV patients, and Mauro et al. (36) used stage I patients.
The quality of the studies was carefully assessed using the NOS, with scores ranging from 7 to 9 (Table 2). Overall, three studies had a score of 9, eight studies had a score of 8, and the other twelve studies had a score of 7, which led to an average score of 7.61. Detailed descriptions of these studies are summarized in Table S3.
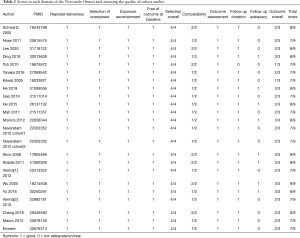
Full table
Prognostic value of ERβ protein expression in different subcellular localizations
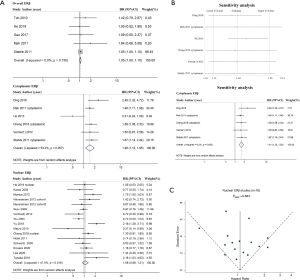
Because four studies reported two HRs based on the different subcellular localizations of ERβ (2,10,32,34), we performed a meta-analysis by different subcellular localizations of ERβ (overall ERβ, cytoplasmic ERβ, and nuclear ERβ) separately to ensure that the HRs of different ERβ subcellular localizations in the same study population were not pooled together (Figure 2). High overall ERβ expression was associated with a poorer OS (HR: 1.05; 95% CI: 1.00 to 1.10, P=0.034) than low overall ERβ expression, and there was no evidence of significant heterogeneity across the studies (I2=0.0%, P=0.730). High cytoplasmic ERβ expression was associated with an even poorer OS than those with low expression, with a pooled HR of 1.48 (95% CI: 1.13 to 1.95, P=0.005), and moderate heterogeneity was indicated using a random-effects model (I2=53.4%, P=0.057). Nuclear ERβ expression was not a predictor of OS, achieving a pooled HR of 1.08 (95% CI: 0.89 to 1.31, P=0.501); moderate heterogeneity was indicated using a random effects model (I2=47.5%, P=0.018).
Subgroup analyses and sources of heterogeneity
Subgroup analyses of different histological types
ERβ is highly expressed in NSCLC and is closely related to the progression of lung adenocarcinoma; therefore, subgroup analysis was performed to investigate potential sources of heterogeneity between studies and assess the consistency of conclusions between lung adenocarcinoma patients and NSCLC patients. We divided all of the studies into two subgroups: studies specific to adenocarcinoma and studies that did not differentiate the histological types of NSCLC.
Among the eight adenocarcinoma studies, five studies included nuclear ERβ (26-29,31,32), two studies included overall ERβ (30,32), and only one study reported cytoplasmic ERβ (25). Therefore, we performed a combined analysis of the three overall ERβ and cytoplasmic ERβ studies (25,30,32) (Figure 3A) and concluded that patients with high expression of overall/cytoplasmic ERβ had statistically longer OS than those with low expression (HR: 1.54, 95% CI: 1.05 to 2.25). Notably, high expression of nuclear ERβ was associated with poor OS in adenocarcinoma patients, with an HR of 1.36 (95% CI: 1.03 to 1.80).
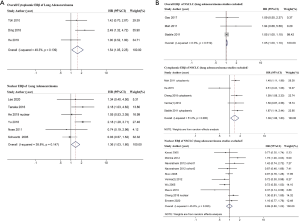
For the 15 studies that did not differentiate the histological types of NSCLC (Figure 3B), high overall ERβ expression and high cytoplasmic ERβ expression were associated with poor OS (HR: 1.05, 95% CI: 1.00 to 1.10 and HR: 1.39, 95% CI: 1.06 to 1.83), and the result indicated no significant association between nuclear ERβ and OS (HR: 0.99, 95% CI: 0.80 to 1.22).
Subgroup analyses of other study characteristics
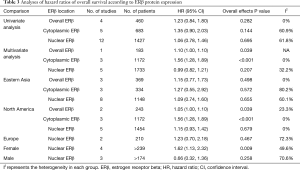
Subgroup analyses were also performed for geographical location and based on whether a multivariate or univariate analysis was used (Table 3). The results indicated that high cytoplasmic ERβ expression was related with a poorer OS than low cytoplasmic ERβ expression in the multivariate analysis group (which was also the North America study group, HR: 1.56, 95% CI: 1.28 to 1.89). These two characteristics may have contributed to the heterogeneity in cytoplasmic ERβ and nuclear ERβ groups because substantial heterogeneity was observed in the univariate analysis and the Eastern Asia and Europe study groups.
Full table
We also pooled sex-specific HRs using data from studies that reported associations separately for men and women (Table 3; Figure S1). The results showed that high nuclear ERβ expression may be associated with a poor OS in women (HR: 1.62, 95% CI: 1.13 to 2.32), and that nuclear ERβ expression had no prognostic value in men (HR: 0.66, 95% CI: 0.32 to 1.36).
Sensitivity analyses
We performed sensitivity analyses using a leave-one-out strategy to evaluate the source of heterogeneity in the cytoplasmic ERβ group (Figure 2B). When we excluded, He (in 2015) et al. (33), the pooled HR was 1.61 (95% CI: 1.35 to 1.92) and the heterogeneity between studies was markedly reduced (I2=0.0%, P=0.692). When we excluded any other single study in turn, the pooled HR of the remaining studies was not substantially altered and ranged from 1.39 (95% CI: 1.06 to 1.83) to 1.47 (95% CI: 1.06 to 2.03) (Figure 2B), indicating that He (2015) et al. (33), which focused on stage IV lung adenocarcinoma only and had a relatively small sample size, may be the main source of heterogeneity in the cytoplasmic ERβ group.
Publication bias of the included studies
We performed publication bias analyses for the nuclear ERβ group (n=16, Figure 2C) and NSCLC nuclear ERβ group (n=10, Figure S2). There was some evidence of publication bias in the inspection of the funnel plot, with two studies reporting standard errors (SEs) greater than those of other studies, however, the Egger’s test was not significant (P=0.883 for nuclear ERβ group and P=0.616 for NSCLC nuclear ERβ group).
Survival analysis of the ERβ mRNA expression in lung adenocarcinoma
Survival analyses were performed with prognostic information from the tumor samples of TCGA-LUAD (n=526), and the mRNA expression of none of the four genes showed statistical prognostic value (Figure 4, Figure S3). The results from the Kaplan-Meier Plotter analysis of 719 patients with lung adenocarcinoma patients indicated a non-significant prognostic value of ERβ mRNA expression (HR: 1.23, 95% CI: 0.98 to 1.55) (Figure 4B). These results indicate that the ERβ mRNA expression may not be a prognostic predictor of lung adenocarcinoma.
Patient characteristics
Among the 39 patients for IHC, 23 (59.0%) were men and 16 (41.0%) were women. The clinicopathological factors of the patients are shown in Table 4. Most of the patients were diagnosed with lung adenocarcinoma (79.5%) and eight patients were diagnosed with squamous cell lung carcinoma (20.5%). All of the patients had stage III disease, of which 27 (69.2%) had stage IIIA disease and 12 (30.8%) had stage IIIB disease.

Full table
Tissue samples for western blotting and RT-qPCR were obtained from a female patient diagnosed with adenocarcinoma and a male patient diagnosed with squamous cell carcinoma.
ERβ expression in tumor and non-tumor tissue
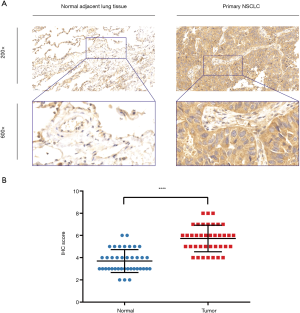
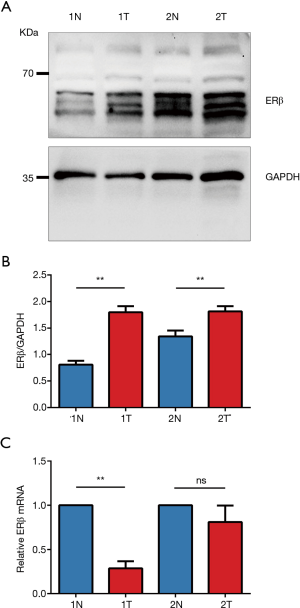
IHC staining showed that 32 patients (82.1%) had high ERβ expression and seven patients (17.9%) had low ERβ expression in the tumor tissues, whereas only nine patients (23.1%) had high ERβ expression in the normal adjacent tissues. The mean ERβ score in the tumor and normal adjacent tissues is 5.72 and 3.69, respectively. Overall, the IHC score of ERβ protein expression was significantly higher in the tumor tissues than in the normal adjacent tissues (P<0.0001) (Figure 5).
In the lung adenocarcinoma gene expression profile dataset, 526 cases were tumor tissues (all from TCGA-LUAD) and 347 cases were normal tissues (288 from GTEx-LUNG and 59 from TCGA-LUAD). ERβ mRNA expression was not significantly different between tumor tissues and normal tissues (Figure 4C,D; Table S4). To verify the differential expression in the above dataset, four GEO datasets (Table S5) of lung adenocarcinoma were selected. The heatmap drawn from the logFC and p-values showed that the relatively stable ERβ mRNA expression (Figure 4E).
To further validate the inconsistency between ERβ protein and mRNA expression, we analyzed the protein and mRNA expression in paired clinical tissues from two patients (Figure 5). Consistent with the IHC results, Western blotting of paired tissues from the two patients (Figure 6) also showed that the protein expression of ERβ was more prominent in the NSCLC tissues than in the normal adjacent tissues (Figure 6A,C). However, the corresponding ERβ mRNA expression of one of the two patients showed no significant difference between the cancerous tissues and the normal adjacent tissues; the other patient showed even lower ERβ mRNA expression in the cancerous tissues than in the normal adjacent tissues (Figure 6B,D).
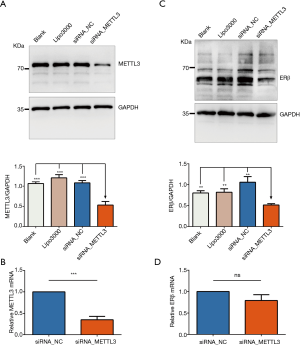
Effect of METTL3 knockdown on ERβ expression
To investigate the possible role of METTL3 in the regulation of ERβ expression, we measured the protein and mRNA levels of ERβ after knockdown of METTL3 in the A549 cell line. The validation of the efficiency of siRNA_METTL3 by RT-qPCR and western blotting is shown in Figure 7. Interestingly, we found that knockdown of METTL3 had a minor effect on ERβ mRNA levels, whereas it resulted in a significant decrease in ERβ protein levels. These results indicated that METTL3 might regulate the translation process of ERβ and cause high ERβ protein expression level in the presence of low ERβ mRNA expression level.
Discussion
Summary of the main results
The present systematic review and meta-analysis summarized data from 23 independent studies to clarify the association between ERβ protein expression and lung cancer survival. We demonstrated that high overall ERβ and cytoplasmic ERβ were negatively associated with the OS of NSCLC, and this association was consistent in lung adenocarcinoma. High nuclear ERβ expression had no effect on NSCLC OS, however, it had a negative effect on OS in lung adenocarcinoma and the female population. Our results conflict with those of two previous meta-analyses (11,12). This conflict may be due to the following reasons. First, we included lung adenocarcinoma-specific studies additional newly published NSCLC studies via a systematic literature search. Second, we excluded studies of ERβ at the mRNA level to ensure that all of the retrieved studies evaluated ERβ protein levels. Third, we extracted additional and detailed prognosis information from each study and performed comprehensive analyses using different subcellular localizations. In addition to survival analysis based on protein level, we performed bioinformatics analysis and found that ERβ mRNA was not predictive of lung adenocarcinoma survival, which is consistent with the conclusion of a meta-analysis published in 2015 (14). Therefore, our results revealed that the prognostic value of ERβ protein and mRNA expression is different.
Public database analysis revealed that ERβ mRNA was not highly expressed in lung adenocarcinoma tissues, when compared with that in normal lung tissues. Our validation in paired tissues of patients who underwent surgical resection also showed that ERβ mRNA was not highly expressed in NSCLC. All this evidence from mRNA level is inconsistent with the high ERβ protein level in tumor tissues proved by our IHC results and western blotting results from paired tissues of patients. To further explore the possible mechanism of this inconsistency, we knocked down METTL3 in an NSCLC cell line and found that protein—but not mRNA—expression of ERβ was downregulated.
We found that ERβ protein was highly expressed in tumor tissues, when compared with that in non-tumor tissues; however, ERβ mRNA was undifferentiated. We propose that ERβ mRNA is regulated by post-transcriptional modifications. Post-transcriptional modification of mRNA regulates protein expression in many tumors (43). For example, Choe et al. reported that mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis (44). Liu et al. reported that m6A methylation regulated CTNNB1 (which encodes β-catenin) to promote the proliferation of hepatoblastoma (45). Arango et al. reported that N4-acetylcytidine (ac4C) is an mRNA modification that improves translation efficiency (46). We propose that the aberrant post-translational modification of m6A mediated by METTL3 in NSCLC tissues enhanced the subsequent translation process, which in turn resulted in the inconsistency between ERβ protein and mRNA levels. Therefore, further studies are required for us to explore the specific mechanism by which METTL3 regulates the expression of ERβ in NSCLC progression.
The downregulation of ERβ mRNA in tumor tissues was reported by Read et al. in 1989, in which the estrogen signaling pathway in MCF-7 cells was activated after estrogen stimulation; however, the mRNA level of ERβ was decreased, which may be a negative feedback regulation (47). Another study reported that, in astrocyte tumors, patients with high E2 levels had low ERα mRNA levels and poor prognosis (48). Therefore, it is possible that the more active the ER signaling pathway, the lower the ER mRNA level.
Further studies are also required for the different effects of nuclear and cytoplasmic ERβ in NSCLC, particularly lung adenocarcinoma. ERβ of different subcellular localizations exerts its influence via genomic and non-genomic signaling. Zhang et al. reported that mitochondrial ERβ may be involved in the inhibition of apoptosis by disrupting the interactions of Bad-Bcl-XL and Bad-Bcl2 in NSCLC (49). Our team reported that ERβ up-regulates the expression of IL-6 to promote the progression of lung adenocarcinoma (7). ERβ and insulin-like growth factor 1 synergistically promote the development of lung adenocarcinoma (17). Nuclear ERβ is a transcription factor in the genomic pathways (50), and it may increase lncRNA-MALAT1 (MALAT1) expression by directly binding to the estrogen response elements located on the promoter of MALAT1 (31), leading to enhanced tumor cell proliferation, invasion, and metastasis and poor prognosis in lung adenocarcinoma patients. However, further research is needed to explain why cytoplasmic ERβ leads to a poorer prognosis than nuclear ERβ.
Limitations
Several limitations of our meta-analysis were noted. The effect sizes in some of the retrieved studies were not available and were estimated from the Kaplan-Meier survival curve. Heterogeneity existed between the study populations of retrieved studies because some studies focused on a female population, high aromatase population or EGFR wild-type population only (29,31,34). Because only five studies mentioned that they used antibodies that only detect ERβ1 and the other studies did not use ERβ isoform-specific antibodies (Table S3), this meta-analysis could not distinguish between the five isoforms of ERβ. Finally, the semi-quantitative IHC method relies on the experience of technicians and presents discrepancies between antibodies and cut-off points (Table S3) (51). These limitations may create doubt on the reliability of the summary estimates; however, our results were generally stable.
Considering the differentiation of histological types for the first time, our meta-analysis provides the latest evidence to systematically evaluate the prognostic value of cytoplasmic and nuclear ERβ in NSCLC. Our bioinformatics analysis provides additional evidence for the limited prognosis data of ERβ mRNA and proposes a new contradiction that deserves further investigation of the expression of ERβ at the protein and mRNA levels. Our experiments confirmed the protein and mRNA expression in clinical samples and improved the reliability of this inconsistency. We placed an objective hypothesis on this inconsistency, and conducted a preliminary investigation in lung cancer cell lines, which made the research highly comprehensive. In future studies, we will explore the relationship between the m6A modification and the estrogen signaling pathway in NSCLC. In addition, the question of whether METTL3 modification can enhance the translational efficiency of ERβ in promoting the progression of NSCLC will be elucidated further.
Conclusions
In conclusion, this study indicated that the high expression of ERβ protein was associated with poor outcomes in NSCLC, particularly lung adenocarcinoma, and that ERβ mRNA expression had no evident effect on lung adenocarcinoma survival. ERβ protein is highly expressed in NSCLC tissues, whereas ERβ mRNA is not. The METTL3-m6A module might regulate the process of ERβ translation and cause NSCLC progression. Our results contribute to the evaluation of prognosis and clinical decisions for NSCLC. Further biological experiments are required to elucidate the specific mechanism underlying the role of m6A modification of ERβ in NSCLC.
Acknowledgments
We thank professor Shiyi Cao from School of Public Health, Tongji Medical College for the support in methodology of meta-analysis, and we thank all the patients and clinical investigators who were involved in the studies and datasets selected in this article.
Funding: This work was supported by the National Natural Science Foundation of China (NSFC) (No. 82072593).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-658
Peer Review File: Available at https://dx.doi.org/10.21037/jtd-21-658
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-658). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics or Institutional Review Board of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, and written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kuiper GG, Enmark E, Pelto-Huikko M, et al. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A 1996;93:5925-30. [Crossref] [PubMed]
- Stabile LP, Dacic S, Land SR, et al. Combined analysis of estrogen receptor beta-1 and progesterone receptor expression identifies lung cancer patients with poor outcome. Clin Cancer Res 2011;17:154-64. [Crossref] [PubMed]
- Tang H, Liao Y, Zhang C, et al. Fulvestrant-mediated inhibition of estrogen receptor signaling slows lung cancer progression. Oncol Res 2014;22:13-20. [Crossref] [PubMed]
- Liu J, Nie Z, Lei Y, et al. The expression of ERβ2, Bcl-xl and Bax in non-small cell lung cancer and associated with prognosis. Int J Clin Exp Pathol 2017;10:10040-6. [PubMed]
- Hsu LH, Chu NM, Kao SH. Estrogen, Estrogen Receptor and Lung Cancer. Int J Mol Sci 2017;18:1713. [Crossref] [PubMed]
- Baik CS, Eaton KD. Estrogen signaling in lung cancer: an opportunity for novel therapy. Cancers (Basel) 2012;4:969-88. [Crossref] [PubMed]
- Huang Q, Zhang Z, Liao Y, et al. 17β-estradiol upregulates IL6 expression through the ERβ pathway to promote lung adenocarcinoma progression. J Exp Clin Cancer Res 2018;37:133. [Crossref] [PubMed]
- Fan S, Liao Y, Qiu W, et al. Estrogen promotes the metastasis of non-small cell lung cancer via estrogen receptor β by upregulation of Toll-like receptor 4 and activation of the myd88/NF-κB/MMP2 pathway. Oncol Rep 2020; Epub ahead of print. [Crossref] [PubMed]
- Wu CT, Chang YL, Shih JY, et al. The significance of estrogen receptor beta in 301 surgically treated non-small cell lung cancers. J Thorac Cardiovasc Surg 2005;130:979-86. [Crossref] [PubMed]
- Cheng TD, Darke AK, Redman MW, et al. Smoking, Sex, and Non-Small Cell Lung Cancer: Steroid Hormone Receptors in Tumor Tissue (S0424). J Natl Cancer Inst 2018;110:734-42. [Crossref] [PubMed]
- Luo Z, Wu R, Jiang Y, et al. Overexpression of estrogen receptor beta is a prognostic marker in non-small cell lung cancer: a meta-analysis. Int J Clin Exp Med 2015;8:8686-97. [PubMed]
- Ma L, Zhan P, Liu Y, et al. Prognostic value of the expression of estrogen receptor β in patients with non-small cell lung cancer: a meta-analysis. Transl Lung Cancer Res 2016;5:202-7. [Crossref] [PubMed]
- Aresti U, Carrera S, Iruarrizaga E, et al. Estrogen receptor 1 gene expression and its combination with estrogen receptor 2 or aromatase expression predicts survival in non-small cell lung cancer. PLoS One 2014;9:e109659 [Crossref] [PubMed]
- Li W, Tse LA, Wang F. Prognostic value of estrogen receptors mRNA expression in non-small cell lung cancer: A systematic review and meta-analysis. Steroids 2015;104:129-36. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. [Crossref] [PubMed]
- Tang H, Liao Y, Xu L, et al. Estrogen and insulin-like growth factor 1 synergistically promote the development of lung adenocarcinoma in mice. Int J Cancer 2013;133:2473-82. [Crossref] [PubMed]
- Liu C, Liao Y, Fan S, et al. G-Protein-Coupled Estrogen Receptor Antagonist G15 Decreases Estrogen-Induced Development of Non-Small Cell Lung Cancer. Oncol Res 2019;27:283-92. [Crossref] [PubMed]
- Fu S, Liu C, Huang Q, et al. Estrogen receptor β1 activation accelerates resistance to epidermal growth factor receptor-tyrosine kinase inhibitors in non-small cell lung cancer. Oncol Rep 2018;39:1313-21. [Crossref] [PubMed]
- Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. J Clin Epidemiol 2021;134:178-89. [Crossref] [PubMed]
- Wells GA SB, O'Connell D. The Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815-34. [Crossref] [PubMed]
- Kawai H, Ishii A, Washiya K, et al. Estrogen receptor alpha and beta are prognostic factors in non-small cell lung cancer. Clin Cancer Res 2005;11:5084-9. [Crossref] [PubMed]
- Nose N, Sugio K, Oyama T, et al. Association between estrogen receptor-beta expression and epidermal growth factor receptor mutation in the postoperative prognosis of adenocarcinoma of the lung. J Clin Oncol 2009;27:411-7. [Crossref] [PubMed]
- Ding X, Li L, Tang C, et al. Cytoplasmic expression of estrogen receptor β may predict poor outcome of EGFR-TKI therapy in metastatic lung adenocarcinoma. Oncol Lett 2018;16:2382-90. [Crossref] [PubMed]
- Lee JH, Kim HK, Shin BK. Expression of female sex hormone receptors and its relation to clinicopathological characteristics and prognosis of lung adenocarcinoma. J Pathol Transl Med 2020;54:103-11. [Crossref] [PubMed]
- Nose N, Uramoto H, Iwata T, et al. Expression of estrogen receptor beta predicts a clinical response and longer progression-free survival after treatment with EGFR-TKI for adenocarcinoma of the lung. Lung Cancer 2011;71:350-5. [Crossref] [PubMed]
- Schwartz AG, Prysak GM, Murphy V, et al. Nuclear estrogen receptor beta in lung cancer: expression and survival differences by sex. Clin Cancer Res 2005;11:7280-7. [Crossref] [PubMed]
- Tanaka K, Shimizu K, Kakegawa S, et al. Prognostic significance of aromatase and estrogen receptor beta expression in EGFR wild-type lung adenocarcinoma. Am J Transl Res 2016;8:81-97. [PubMed]
- Toh CK, Ahmad B, Soong R, et al. Correlation between epidermal growth factor receptor mutations and expression of female hormone receptors in East-Asian lung adenocarcinomas. J Thorac Oncol 2010;5:17-22. [Crossref] [PubMed]
- Yu W, Ding J, He M, et al. Estrogen receptor β promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene 2019;38:1225-38. [Crossref] [PubMed]
- He C, He Y, Luo H, et al. Cytoplasmic ERβ1 expression is associated with survival of patients with Stage IV lung adenocarcinoma and an EGFR mutation at exon 21 L858R subsequent to treatment with EGFR-TKIs. Oncol Lett 2019;18:792-803. [Crossref] [PubMed]
- He YF, Luo HQ, Wang W, et al. Clinical features and prognosis-associated factors of non-small cell lung cancer exhibiting symptoms of bone metastasis at the time of diagnosis. Oncol Lett 2015;9:2706-12. [Crossref] [PubMed]
- Mah V, Marquez D, Alavi M, et al. Expression levels of estrogen receptor beta in conjunction with aromatase predict survival in non-small cell lung cancer. Lung Cancer 2011;74:318-25. [Crossref] [PubMed]
- Verma MK, Miki Y, Abe K, et al. Cytoplasmic estrogen receptor β as a potential marker in human non-small cell lung carcinoma. Expert Opin Ther Targets 2012;16:S91-102. [Crossref] [PubMed]
- Mauro LV, Dalurzo M, Carlini MJ, et al. Estrogen receptor β and epidermal growth factor receptor as early-stage prognostic biomarkers of non-small cell lung cancer. Oncol Rep 2010;24:1331-8. [PubMed]
- Monica V, Longo M, Felice B, et al. Role of hormone receptor expression in patients with advanced-stage lung cancer treated with chemotherapy. Clin Lung Cancer 2012;13:416-23. [Crossref] [PubMed]
- Navaratnam S, Skliris G, Qing G, et al. Differential role of estrogen receptor beta in early versus metastatic non-small cell lung cancer. Horm Cancer 2012;3:93-100. [Crossref] [PubMed]
- Skov BG, Fischer BM, Pappot H. Oestrogen receptor beta over expression in males with non-small cell lung cancer is associated with better survival. Lung Cancer 2008;59:88-94. [Crossref] [PubMed]
- Verma MK, Miki Y, Abe K, et al. Co-expression of estrogen receptor beta and aromatase in Japanese lung cancer patients: gender-dependent clinical outcome. Life Sci 2012;91:800-8. [Crossref] [PubMed]
- Gao X, Cai Y, Wang Z, et al. Estrogen receptors promote NSCLC progression by modulating the membrane receptor signaling network: a systems biology perspective. J Transl Med 2019;17:308. [Crossref] [PubMed]
- Enwere EK, Dean ML, Li H, et al. The prevalence and prognostic significance of estrogen receptor beta expression in non-small cell lung cancer. Transl Lung Cancer Res 2020;9:496-506. [Crossref] [PubMed]
- Zhang C, Huang S, Zhuang H, et al. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 2020;39:4507-18. [Crossref] [PubMed]
- Choe J, Lin S, Zhang W, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018;561:556-60. [Crossref] [PubMed]
- Liu L, Wang J, Sun G, et al. m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol Cancer 2019;18:188. [Crossref] [PubMed]
- Arango D, Sturgill D, Alhusaini N, et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 2018;175:1872-1886.e24. [Crossref] [PubMed]
- Read LD, Greene GL, Katzenellenbogen BS. Regulation of estrogen receptor messenger ribonucleic acid and protein levels in human breast cancer cell lines by sex steroid hormones, their antagonists, and growth factors. Mol Endocrinol 1989;3:295-304. [Crossref] [PubMed]
- Dueñas Jiménez JM, Candanedo Arellano A, Santerre A, et al. Aromatase and estrogen receptor alpha mRNA expression as prognostic biomarkers in patients with astrocytomas. J Neurooncol 2014;119:275-84. [Crossref] [PubMed]
- Zhang G, Yanamala N, Lathrop KL, et al. Ligand-independent antiapoptotic function of estrogen receptor-beta in lung cancer cells. Mol Endocrinol 2010;24:1737-47. [Crossref] [PubMed]
- Hershberger PA, Stabile LP, Kanterewicz B, et al. Estrogen receptor beta (ERbeta) subtype-specific ligands increase transcription, p44/p42 mitogen activated protein kinase (MAPK) activation and growth in human non-small cell lung cancer cells. J Steroid Biochem Mol Biol 2009;116:102-9. [Crossref] [PubMed]
- Atmaca A, Al-Batran SE, Wirtz RM, et al. The validation of estrogen receptor 1 mRNA expression as a predictor of outcome in patients with metastatic non-small cell lung cancer. Int J Cancer 2014;134:2314-21. [Crossref] [PubMed]


