
Artificial intelligence-assisted decision making for prognosis and drug efficacy prediction in lung cancer patients: a narrative review
Introduction
Lung cancer is one of the most prevalent malignant neoplasms with highest mortality (1). According to its histological classification, lung cancer consists of small-cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) (1). SCLC, accounting for around 13–15% of lung cancer, contributes to an extremely poor prognosis for patients, with approximate 5-year overall survival (OS) of 5% for its extensive stage (2). NSCLC occupies most of the lung cancer presenting better prognosis than SCLC, which can be divided into lung adenocarcinoma (LUAD), large cell lung cancer (LCLC) and lung squamous cell carcinoma (LUSC) (3). Although the rapid development of treatment for lung cancer such as targeted therapy and immunotherapy has extremely improved the prognosis of NSCLC, its long-term survival rate is still poor (4-6). Therefore, to improve treatment effect of lung cancer, comprehensive usage of multi-dimensional data including image data, clinical symptoms, and laboratory indexes is essential for early diagnosis and developing personalized treatment (7,8). However, it’s difficult for current manual recognition to attain the efficient and accurate clinical decision-making with so much information.
Artificial intelligence (AI) is a frontier technology adopting multiple computer algorithms to comprehensively analyze the imaging and clinicopathological data of patients for early identification of malignant nodules and decision-making assistance, which has achieved higher accuracy and efficiency than manual identification (9). Though the AI technique has rarely been applied to clinical decision-making of lung cancer, recent surge in AI algorithms has showed their potential to accurately screen malignant nodules, predict prognosis and therapeutic effect of lung cancer (10-12), indicating the potential of AI-assisted decision-making for diagnosis, prognosis, and drug efficacy prediction.
We present the following article in accordance with the Narrative Review reporting checklist (available at: https://dx.doi.org/10.21037/jtd-21-864).
Methods
Here, in order to offer an overview of potential for AI in lung cancer clinical decision-making, we searched literature from 2000 through July 31th, 2021 in MEDLINE/PubMed, the Web of Science, the Cochrane Library, ACM Digital Library, INSPEC and EMBASE. Search terms included “artificial intelligence”, “AI”, “deep learning”, “lung cancer”, “NSCLC”, “SCLC” and their combinations. These literatures were then selected by two independent authors. Articles chosen by only one author will be examined by another author to determine whether this article was relative and valuable. Additionally, other relative articles of the selected article authors and the bibliography of selected articles were also reviewed to retrieve all relevant studies. The selected literatures were read by all authors and discussed to draw reliable conclusions. Based on the above work, we reviewed the state-of-the-art application of AI in clinical decision-making and discussed the promise of AI in the field of lung cancer through highlighting the applications of AI in diagnosis, prognosis, and drug efficacy prediction (Figure 1).
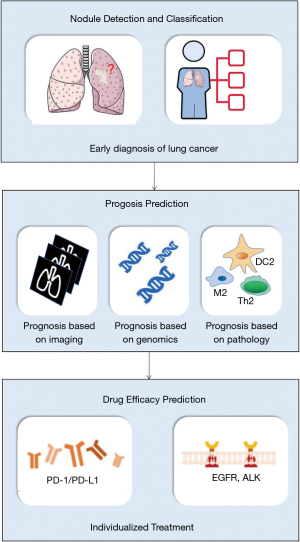
Discussion
Radiomics and deep learning
Radiomics and deep learning are most studied AI technology in the field of medicine (13,14). Radiomic methods are applied to use image-based radiomic features to establish decision-making models for diagnosis, prognosis, drug efficacy prediction and so on (15). Based on numerous data generated and stored in computer and facilitated by the development in algorithms, the radiomic methods are applied to extract traditional image features, including statistical, model-based, transform-based, and shape-based features, to develop the models and assist the decision making for physicians (15). The basic workflow of this method contains five phases: data selection, medical imaging, feature extraction, exploratory analysis, and modelling (16).
Deep learning directly uses the convolutional neural network (CNN), the artificial neural network (ANN), the recurrent neural network (RNN) to extract features, then combines with the full-connection layer to complete classification and prediction (17). Compared with the traditional radiomics approaches, deep learning allows for the automated feature extraction in an incremental manner, and thus they require little human input (Figure 2). Current studies mainly focus on improving the algorithm and strategy of these two steps or combining them to construct a complete computer-aided diagnosis (CADx) system for lung cancer risk prediction. Winkels et al., for instance, proposed a 3D CNN with group convolutions (3D-G-CNNs) method to accurately classify pulmonary nodule. They trained the 3D-G-CNNs model in a relatively small dataset yet yield similar performance to regular CNN model trained on 10 times more data. Their work significantly improved the data efficacy of CNN and provided a potential solution to the problem of insufficient labeled data in training a CNN model with satisfying performance (17). To stress, other solutions to solve the problem of relatively small datasets include data argumentation (18), transfer learning (19), extracting patches on multiple planar view (20,21) and ensemble learning (22), etc.
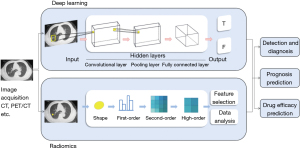
Although radiomics and deep learning have been widely used for decision making of many diseases, such as pneumonia and several tumors, lung cancer remains the most extensively studied field among them (23). By mining large databases and adopting deep learning, biopsy trauma of lung cancer patients can be avoided to the maximum.
Artificial intelligence in nodule detection and classification
One of the major reasons of low long-term survival rate for lung cancer is dissatisfactory early diagnosis. Prospective randomized controlled trails have showed that regular CT screening and early diagnosis can significantly reduce mortality (24,25). AI-based model for lung cancer diagnosis is a decades-old concept, but still promising due to the advances in deep learning algorithms and large-scale databases. Up to now, researchers have developed a large number of models in this field, and even put it into practical use (26,27).
The AI-assisted diagnosis of lung cancer patients typically involved pulmonary nodule detection and classification (Figure 2). In 2019, researches from Google AI proposed an end-to-end deep learning approach to predict the risk of lung cancer. Their approach included three CNN models to perform analysis of whole-CT volumes, detection of the region-of-interest (ROI) and prediction of the risk, intending to replicate radiologist’s workflow. The area under the curve (AUC) reached 94.4% and 95.5% in the test and independent validation set, claiming a similar or even better performance than the radiologist (10). Though it is one of the largest cohort studies, and the model yielded relatively satisfying performance, further validation and more convincing comparison between AI-based models and radiologists need to be conducted. Additionally, such a deep learning model based on the “black box” cannot guarantee that existing diagnostic guidelines will always be followed. In such a context, new clinical guidelines may be established, however, it will take some time before the new guidelines are accepted by radiologists (28,29).
Artificial intelligence in prognosis (Table 1)
Table 1
| Author | Year | Method | Dataset | Train cohort | Validation cohort | Test cohort | Model | Outcome | Performance reported |
|---|---|---|---|---|---|---|---|---|---|
| Coroller TP | 2017 | Retrospective multi-center on CT images | 85 NSCLC | 13 | NA | 72 | Radiomic mapping | 3-year overall survival | AUC: 0.65 in the pathological complete response cohort; 0.73 in the gross residual disease cohort |
| Hosny A | 2018 | Retrospective multi-center on CT images | 1,162 NSCLC | 517 | 237 | 408 | CNN | 2-year overall survival | AUC: 0.70 in the radiotherapy cohort; 0.71 in the surgery cohort |
| Xu Y | 2019 | Retrospective multi-center on CT images | 268 NSCLC | 107 | NA | 161 | CNN | 2-year overall survival and mortality risk | AUC (2-year overall survival): up to 0.74; HR (mortality risk): 6.16 |
| Kim H | 2020 | Retrospective multi-center on CT images | 908 LUAD | 800 | 800 internal 108 external | NA | CNN | Disease-free survival | HR: 2.5 in the internal validation; 3.6 in the external validation |
| Wang X | 2020 | Retrospective single-center on CT images | 173 NSCLC | 124 | Cross validation | 49 | Radiomic mapping | 3-year overall survival | AUC: 0.92 in the cross validation; 0.84 in the test cohort |
| Li YY | 2018 | Retrospective study on gene expression | 502 LUAD | 336 | NA | 166 | univariate Cox regression | 3-year overall survival | AUC: 0.752 and 0.705 in the training and test cohorts |
| Li Y | 2019 | Retrospective study on gene expression | 1,071 LUAD | 492 | 347 | 232 | sigFeature, random forest, and univariate Cox regression | 5-year overall survival | AUC: 0.656, 0.753, and 0.739 in the training, validation, and test cohorts |
| Luo X | 2017 | Retrospective multi-center on H&E images | 1,034 NSCLC | 523 | NA | 511 | Cox proportional hazards analysis and random survival forest | Overall survival | HR: 2.34, 2.22 in training and test cohorts |
| Corredor G | 2019 | Retrospective multi-center on H&E images | 301 NSCLC | 70 | Set 1: 119 Set 2: 112 |
231 | Watershed-based algorithm and QDA classifier | Recurrence-free survival | HR: 2.80, 4.45, and 3.08 in the set 1, set 2 and test cohorts |
CT, computed tomography; NSCLC, non-small cell lung cancer; AUC, area under the curve; CNN, convolutional neural network; LUAD, lung adenocarcinoma; NA, not available; HR, hazard ratio; QDA, quadratic discriminant analysis.
Prognosis based on imaging
Despite numerous advancements in lung cancer treatment over the last decades, the five-year survival rates of lung cancer patients are still relatively low (overall 19%) (30). The epidemiological statistics have demonstrated a mortality of 75% because of the inaccurate prognosis analysis and impropriate treatments (31). Therefore, the accurate prediction of clinical outcomes is crucial to increase cure rate and survival rate for lung cancer patients.
Imaging is an important technology used to aid in lung cancer diagnosis and evaluation (32). Furthermore, it has been proved by many studies over the past two decades that images contain much accessible information. On this basis, the imaging technology shows a potential role in the context of personalized precision medicine (29). Recently, many researchers have focused on the correlation between radiomics and lung cancer prognosis, and several radiomics signatures have been developed to assist decision making for prognosis (31,33,34). Imaging modality for most radiomics signatures are CT scans. For example, Huang et al. developed a radiomics signature to estimate disease-free survival (DFS) in patients with early-stage NSCLC. The signature based on 282 patients was generated by the least absolute shrinkage and selection operator (LASSO) COX regression model. The signature demonstrated a better performance (P<0.0001) for the estimation of DFS (C-index: 0.72) than the clinical-pathologic nomogram, as well as a more accurate classification of survival outcomes (35). Furthermore, studies on deep learning have demonstrated strong potential of prognostic analysis. In 2018, Hosny et al. designed a prognostic signature through CT data, which used a 3D CNN to predict the 2-year-survival of patients treated with radiotherapy and surgery. Their study showed that the CNN predictions were significantly associated with 2-year OS from the start of respective treatment for radiotherapy (AUC =0.70) and surgery (AUC =0.71) patients (36). Incorporating CT at different time point is another strategy to integrate more information, thus to achieve more accurate prognosis. Xu et al. built a prognostic model for NSCLC patients based on CNN with time encompassing RNN which can combine pre-treatment and post-treatment CT-images at 1, 3 and 6 months. By adding more CT-images at different time points, they observed a significant increase of performance (only pretreatment CT: AUC =0.58, P=0.3, Wilcoxon’s test; add 1 mouth CT AUC =0.64, P=0.04; add 3 months CT: AUC =0.69, P=0.007; add 6 months CT: AUC =0.74, P=0.001), indicating the great potential of multi-time point strategy (37).
Apart from traditional feature engineering approaches, imaging features combining clinical models have played an important role in prognosis analysis. However, whether the output of the deep learning model is an independent risk factor for prognosis needs to be evaluated. Therefore, Kim et al. developed a CT-based deep learning survival prediction model (DLPM) to predict DFS of LUAD patients and validated it with other prognostic factors. They generated a Cox regression model combining the DLPM outputs and other clinical factors [C-index, 0.71; 95% confident interval (CI): 0.61–0.80]. The hazard ratio (HR) of the DLPM outputs were 2.5 (95% CI, 1.03–5.9) in the internal validation and 3.6 (95% CI: 1.6–8.5) in the external validation, which indicated that they can serve as a powerful independent prognostic factor (38). Meanwhile, Wang et al. combined radiomic features of CT and clinical data to build a mixed prognostic analysis model, reaching an accuracy of 88.7% (AUC =0.92) on an independent data set and 79.6% on the other independent data set (31). The accuracy of their models is relatively high due to the combination of imaging features and other clinical data, indicating that the multi-omics approach may be a breakthrough for the application of AI models in clinical prognosis of lung cancer patient.
Although a large number of prognostic models based on images have been developed and reported in the past few years, these models still have many defects in practices of clinical application. For example, some of them only included a small sample size and lacked external validation (35). Although some other studies adopted external validation, the sample of which merely involved patients with similar distribution (36,38), which limits its application for patients in other countries or continents. Thus, further studies with large sample size from various races should be considered to develop and validate novel models to make them truly satisfy the clinical application.
Prognosis based on genomics
Apart from features of imaging to assist decision making for prognosis, it has been long proved that genome can serve as prognostic factors for lung cancer (39-41). Because of the establishment of The Cancer Genome Atlas (TCGA) and other large-scale public databases providing patients information including genomic and clinical information, researchers can share and obtain data to analyze the relation between cancer and genomic information and develop diagnostic or prognostic models (42,43). Collecting and analyzing data from TCGA, Yu et al. established a Support Vector Machine (SVM) model to predict the 3-year-survival of LUAD patients. When combining 85 significant genes, the model achieved 0.896 for AUC of receiver operating characteristic curve (ROC), relatively satisfying for this problem. Additionally, their 28-gene and 7-gene model achieved 0.810, 0.711 for AUC of ROC, respectively. However, the performance of this model is yet to be validated in independent datasets containing more patients at different stages with different lung cancer subtypes (44). Li et al. collected data form both TCGA and Gene Expression Omnibus (GEO) and selected survival related genes by a novel combination strategy of integrating three different algorithms (sigFeature, random forest, and univariate Cox regression). After the selection, 892 candidate genes were further evaluated by LASSO Cox regression analysis. After 100,000 times of calculation and model construction, they generated a prognostic model based on 16 genes to classify LUAD patients into high- and low-risk groups, reaching 0.753, 0.726 and 0.656 for AUC of ROC at 1-, 3- and 5-year OS in the TCGA cohort and 0.822, 0.714, 0.753 in the external validation set. Eventually, they built a nomogram based on their model outputs and other clinical factors to predict individual prognosis, the C-index for 1-, 3- and 5-year OS reached 0.695, 0.694 and 0.695 respectively (45).
Although genomic information gives a new idea of developing prognosis prediction models, accuracy of these models has not worked out as intended, thus researchers set their sights on combining clinical data and heterogeneously expressed gene (46-48). For example, Lai et al. combined seven well-known biomarkers and eight differentially expressed gene biomarkers based on microarray data with clinical information to build a deep neural network, reaching 0.8163 for AUC of ROC in predicting the 5-year survival of NSCLC patients (47), which is significantly higher than the models above in the 5-year survival prediction. Furthermore, other researchers developed binned time survival analysis (DeepBTS) models to select 14 features for prognosis prediction, which avoided manually assuming proportional hazards and approximating survival data of Cox proportional hazard model (46). In the future, DeepBTS has the potential to combine imaging information, genomic and clinicopathologic data to improve its performance.
Hopefully, with the development of large-scale cancer database, the improvement of algorithm and growing understanding of cancer related genes, the prognosis models based on genomics can be applied to the clinical practice, thus to assist decision-making in lung cancer prognosis and patient management.
Prognosis based on pathology
Pathological features of lung cancer samples, especially in whole slide images (WSIs) contain a vast amount of prognostic information to be excavated. Most research institutions prefer digital slides to glass slides and a conventional light microscope, because of the information displayed in the two sets of slides is highly consistent and digital slides can scan and store efficiently and accurately with little use of laboratory personnel (49-51). In 2016, Yu et al. demonstrated the potential of deep learning model to predict the survival outcomes of LUAD and LUSC patients. Extracting 9,879 quantitative features of 2,480 haematoxylin/eosin stained WSIs by an automatic image segmentation PEPline and evaluating these features with 7 distinct classifiers, they selected 80 candidates features by three classifiers achieving 0.83 for AUC of ROC. In order to predict the survival outcomes more efficiently, they built elastic net-Cox hazards models to reduce the numbers of features. In independent data set, the model can distinguish LUAD patients with shorter and longer survival time (P=0.028, log-rank test) and LUSC patients (P=0.035, log-rank test) (11).
Apart pathological features extracted and selected from the whole slide as prognostic factors, more and more studies have focused on specific pattern of cancer histopathology such as tumor-infiltrating lymphocytes (TILs) (49-51). Integrating the density, co-localization and spatial architecture of the TIL clusters, Corredor et al. trained a classifier to predict the recurrence in early-stage NSCLC. The TIL features were examined to be strongly associated with likelihood of recurrence (log rank P<0.02) and a multivariate Cox Proportional Hazards analysis revealed a strong predictive power of the developed classifier (HR: 3.08, 95% CI: 2.1~4.5, P=7.3×10−5) outperforming two expert thoracic pathologists (51).
Represented by the researches above, a surge of studies in AI-assisted prognosis field have been witnessed in the last few years with the development of computer science and medical science such as deep learning, radiomics, genomics (Table 1). Despite the demonstrated relatively high performance of the recently established models, few have already been put into clinical use due to the complication of individual variation in the prognosis of lung cancer patients. Predictably, in the near future, by combining with other disciplines AI may reach to its full potential to integrate an accurate and comprehensive prognostic model with robust performance for oncology.
Artificial intelligence in drug efficacy prediction
Efficacy prediction of immunotherapy
Immunotherapy has shown remarkable success in the treatment of lung cancer and other malignant tumors. Immune checkpoints inhibitors (ICIs) such as antibodies directed to the programmed cell death protein 1/programmed cell death 1 ligand 1 (PD-1/PD-L1) have reached clinic after European Medicines Agency and Food and Drug Administration approved (52-54). Though the overall efficacy of these drugs is relatively high comparing to traditional chemotherapeutic drug, the response rate is still unsatisfying within the none-selected patients (19% with nivolumab) (55). It is essential to accurately classify patients with their potential response to immunotherapy to avoid the unnecessary expensive administration of these drugs and potential toxic effects (Table 2).
Table 2
| Author | Year | Method | Dataset | Train cohort | Validation cohort | Test cohort | Model | Outcome | Performance reported |
|---|---|---|---|---|---|---|---|---|---|
| Khorrami M | 2020 | Retrospective multi-center on CT images | 139 NSCLC | 50 | Set 1: 62 Set 2: 27 |
NA | Radiomic mapping | Response to immunotherapy | AUC: 0.88 in the training cohort; 0.85 and 0.81 in the validation cohort |
| Tian P | 2021 | Retrospective single-center on CT images | 939 NSCLC | 750 | 93 | 96 | 3D CNN DenseNet121 | PD-L1 expression Treatment response | AUC: 0.78, 0.71, and 0.76 in the training, validation, and test cohorts |
| Rios Velazquez E | 2017 | Retrospective multi-center on CT images | 763 NSCLC | 353 | 352 | NA | Radiomic mapping plus clinical models | EGFR and KRAS mutation | AUC =0.75 in EGFR(+)/EGFR(-), AUC =0.69 in KRAS(+)/KRAS(-), AUC =0.86 in EGFR(+)/KRAS(+) |
| Wang S | 2019 | Retrospective multi-center on CT images | 844 LUAD | 603 | 241 | NA | CNN | EGFR mutation | AUC: 0.85 and 0.81 in the training and validation cohorts |
| Song J | 2018 | Retrospective multi-center on CT images | 314 NSCLC | 117 | Set 1: 101 Set 2: 96 |
NA | Radiomic mapping | PFS of EGFR-TKI therapy | HR: 3.61 in the training cohort; 3.77 and 3.67 in the validation cohorts |
| Mu W | 2020 | Retrospective multi-center on PET/CT images | 681 NSCLC | 429 | 187 | 65 | 2D SResCNN model | EGFR mutation Treatment response | AUC: 0.86, 0.83, and 0.81 in the training, validation, and test cohorts |
| Song J | 2020 | Retrospective multi-center on CT images | 342 NSCLC | 145 | Set 1: 101 Set 2: 96 |
NA | Radiomic mapping | PFS of EGFR-TKI therapy | HR: 2.13 in the training cohort; 2.35 and 2.32 in the different validation cohorts |
| Dercle L | 2020 | Retrospective multi-center on CT images | 188 NSCLC | 135 | 53 | NA | Radiomic mapping | Treatment response to nivolumab, docetaxel, and gefitinib | AUC: nivolumab, 0.77; docetaxel, 0.67; gefitinib, 0.82 |
| Song Z | 2021 | Retrospective multi-center on CT images | 1,028 NSCLC | 651 | 286 | 91 | 3D CNN ResNet10 | ALK fusion status Treatment response | AUC (CNN): 0.8046, 0.7754 in the primary and validation cohorts AUC (trained by both CT images and clinicopathological information): 0.8540, 0.8481 in the primary and validation cohorts |
CT, computed tomography; NSCLC, non-small cell lung cancer; AUC, aera under the curve; CNN, convolutional neural network; LUAD, lung adenocarcinoma; NA, not available; HR, hazard ratio; PFS, progression-free survival; PET, positron emission tomography; PD-L1, programmed cell death 1 ligand 1; ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitors; SResCNN, small-residual-convolutional-network.
Radiomics allows quantitatively extracting features of the images in a high throughput manner and constructs a prediction model by evaluation and integration of these features (Figure 2). Strategies based on radiomics reflect full information of the whole tumor in a non-invasive way to predict individual response to immunotherapy compared with the traditional biopsy-based assays which neglect the heterogeneity of the tumor (56,57). On the basis of the correlation between tumoral/peritumoral immune infiltration and patient response to PD-1 and PD-L1 immunotherapy, early study in 2018 demonstrated a radiomic signature of tumoral/peritumoral immune infiltration. They reported that the signatures of CT images could discriminate inflamed tumors from immune-desert tumors (AUC =0.76). In addition, they generated a radiomic score which was positively associated with patients’ response at 3 months, 6 months and overall survival (58). In 2019, based on the preprocessing of the basic features by radiomics to enhanced CT, a non-invasive machine-learning marker was developed to identify immunotherapy responses and non-responses in the anti-PD-1 therapy of melanoma and NSCLC. They observed significant performances in OS prediction for both tumor types (NSCLC: 0.76= AUC, P<0.01; melanoma: 0.77= AUC, P<0.01), with a significant survival difference at 1-year of 25%. However, this radiomics immunotherapy response biomarker has no obvious effect on predicting overall survival in patients treated with cytotoxic chemotherapy (P=0.07) (59). Another study took Response Evaluation Criteria in Solid Tumors (RECIST) as the benchmark and analyzed changes in the intertumoral and peritumoral texture patterns of lung CT to predict patients’ response to ICIs. Their classifier was able to distinguish between patients responding to ICIs and patients nonresponding to ICIs with an AUC of 0.88. To further estimate their selected feathers, they also generated a risk-score system which was associated with overall survival (HR: 1.64, C-Index =0.72). Interestingly, they found that their selected peritumoral features correlated with the TIL density, which can corroborate with the study of Sun et al. mutually (58,60).
Previous studies mainly focus on predicting efficacy of immunotherapy by radiomics. As the research continues, the combination of radiomics and deep learning may be a state-of-art strategy (Figure 2). Recently, Tian and his colleagues developed a PD-L1 expression signature (PD-L1ES) based on CT images of 939 NSCLC patients, a relatively large dataset. By integrating radiomics and deep learning features, the signature demonstrated its potential to predict high PD-L1 expression (PD-L1 ≥50%) patients, thus to select responsive patients to PD-1 and PD-L1 immunotherapy (AUC =0.78, 0.71, 0.76 in training, validation and test cohorts). To estimate the effectiveness of this strategy, they confirmed the features extracted by deep learning and radiomics were complementary. Furthermore, an additional combination of a clinical model can significantly improve the stratification capabilities of the signature (HR: 3.53, 95% CI: 1.86–6.72; P<0.001) (61).
Efficacy prediction of targeted therapy
Targeted therapy is another revolutionary progress that significantly improved outcomes of lung cancer patients in recent years. To identify responsive patients to targeted therapy, identification of predictive biomarkers such as epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) are of great importance (40,62,63). Apart from traditional and invasive biopsy, a growing body of studies tends to establish non-invasive AI-based models to predict the oncogene mutations and clinical outcomes of lung cancer patients (64-68). For example, Wang et al. proposed an end-to-end deep learning model by learning EGFR mutation-related features from 14,926 CT images of 844 patients to predict the EGFR-mutant probability. Their model yelled on encouraging results, both in the primary cohort (AUC =0.85, 95% CI: 0.83–0.88) and independent validation cohort (AUC =0.81, 95% CI: 0.79–0.83) (68). Similarly, Mu et al. developed a deep learning model based on both CT and 18F-FDG-PET images of NSCLC patients. The model output an EGFR deep learning score (EGFR-DLS) which was positively associated with PFS in patients treated with EGFR- tyrosine kinase inhibitors (TKIs). Combining with the clinical features, their model achieved AUCs of 0.88, 0.88, and 0.84 for ROC in the training, internal validation, and external test cohorts, indicating potential application as a tool to predict the clinical outcomes of patients treated with TKIs and ICIs (67).
Another end-to-end deep learning model developed by Song et al. predicted the efficacy of EGFR-TKI therapy to stage IV EGFR variant–positive NSCLC patients by progression free survival (PFS). Their prognostic signature based on LASSO Cox proportional hazards regression were able to distinguish between patients with low risk of progression and high risk of progression after receiving EGFR-TKI therapy (HR =2.13, 95% CI: 1.30–3.49). Furthermore, they discovered that high-progression-risk patients have no significant difference with patients receiving first-line chemotherapy in PFS, indicating EGFR-TKI therapy was less beneficial to these patients (65).
Apart from EGFR-TKI, efficacy prediction of other targeted therapy was also investigated recently. Song et al. trained a deep learning model by both CT images and clinicopathological information to predict ALK fusion status in NSCLC patients. The performance of the model only based on CT images achieved 0.8046 (95% CI: 0.7715–0.8378) and 0.7754 (95% CI: 0.7199–0.8310) for AUC in the primary and validation cohorts respectively. However, the AUC of their model raised to 0.8540 (95% CI: 0.8257–0.8823) and 0.8481 (95% CI: 0.8036–0.8926) by combining the clinicopathological information (64).
These established signature and models have revealed the potential of AI in drug efficacy prediction and their strategies may provide reference for future studies (Table 2). As modern medicine further elucidating molecular mechanisms of tumor and more new therapies being developed, AI-based models for drug efficacy prediction may strive to answer the eager need for personalized treatment and precision medicine.
Conclusion and future challenges
As an indispensable approach to precision medicine, artificial intelligence is capable of assisting clinical decision making from many aspects. In this review, we summarized three important applications of AI-assisted decision making in lung cancer patients and presented some frontier studies in these field. These studies demonstrated the advantages of AI, especially of deep learning and radiomics, for its quantitatively interpretation of patients’ information and its potential to deal with the dynamics, individual differences and heterogeneity of lung cancer. Utilizing radiomics and deep learning for decision making could have achieved promising efficiency. In terms of nodule detection and classification, some deep learning approaches even exerted better performance than physicians, whose AUC achieved 0.955 in the validation set (10), which was close to biopsy. Moreover, some algorithms for prognosis and drug efficiency prediction showed the AUC over 0.90 in survival prediction and AUC over 0.85 in gene mutation evaluation (31,60), which provided non-invasive methods to precisely evaluate status of mutant gene and overall survival. In summary, AI technology has the potential to completely replace classic methods based upon histopathological examination and biopsy through continuously improving the accuracy of the prediction model.
However, challenges are still to be solved in many aspects. First, the current models mostly depend on retrospective study, which inevitably caused biases such as selection bias and information bias (68,69). Therefore, prospective studies are urgently needed to verify the efficiency of these model. Second, generalizability of models has certain limitations. Some studies only included small sample size for validation, making it difficult to evaluate the generalizability of algorithms (35). Other researches involved lager sample size, but patients of their external validation have similarly geographic distribution (38). Thus, multi-center studies with large samples from different regions are still needed to verify the efficiency of the existing models. Third, for drug efficiency prediction, the models of current studies were based on previously approved clinical drugs (64,69). Nevertheless, whether they are suitable for drug efficiency prediction of the novel inhibitors such as Alectinib and Ceritinib is uncertain, so it is necessary to include data of these drugs to validate the stability of the models or even develop better algorithms. Furthermore, for survival prediction, the existence of censored data and incomplete OS make it uncertain whether these models have predictive value in predicting prognosis (11,37). In the future, these researches should include both retrospective and prospective data to further verify and optimize the algorithm.
In summary, AI gradually shows the potential as a smart assistant for radiologists and oncologists to detect lung cancer and develop personalized treatment plan. However, the limited sample quantity and quality impede the clinical practice of radiomics and deep learning. Future multi-center studies should adopt large prospective data from various countries and continents to further optimize and validate existing models.
Acknowledgments
Funding: This research was supported by National Guided Science and Technology Development Project of Sichuan Province (2020ZYD005); the Science and Technology Project of Chengdu (2017-CY02-00030-GX); the Science and Technology Project of Sichuan (2020YFG0473); Postdoctoral Program of Sichuan University (2021SCU12018) and Postdoctoral Program of West China Hospital, Sichuan University (2020HXBH084).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Jianxing He and Hengrui Liang) for the series “Artificial Intelligence in Thoracic Disease: From Bench to Bed” published in Journal of Thoracic Disease. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at: https://dx.doi.org/10.21037/jtd-21-864
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-864). The series “Artificial Intelligence in Thoracic Disease: From Bench to Bed” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang C, Tan S, Li J, et al. CircRNAs in lung cancer - Biogenesis, function and clinical implication. Cancer Lett 2020;492:106-15. [Crossref] [PubMed]
- Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346-60.e7. [Crossref] [PubMed]
- Lewis DR, Check DP, Caporaso NE, et al. US lung cancer trends by histologic type. Cancer 2014;120:2883-92. [Crossref] [PubMed]
- Doroshow DB, Bhalla S, Beasley MB, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol 2021;18:345-62. [Crossref] [PubMed]
- Dovedi SJ, Elder MJ, Yang C, et al. Design and efficacy of a monovalent bispecific PD-1/CTLA4 antibody that enhances CTLA4 blockade on PD-1+ activated T cells. Cancer Discov 2021;11:1100-17. [Crossref] [PubMed]
- Garassino MC, Martelli O, Broggini M, et al. Erlotinib versus docetaxel as second-line treatment of patients with advanced non-small-cell lung cancer and wild-type EGFR tumours (TAILOR): a randomised controlled trial. Lancet Oncol 2013;14:981-8. [Crossref] [PubMed]
- Huynh E, Hosny A, Guthier C, et al. Artificial intelligence in radiation oncology. Nat Rev Clin Oncol 2020;17:771-81. [Crossref] [PubMed]
- Murphy SJ, Harris FR, Kosari F, et al. Using genomics to differentiate multiple primaries from metastatic lung cancer. J Thorac Oncol 2019;14:1567-82. [Crossref] [PubMed]
- Liu A, Wang Z, Yang Y, et al. Preoperative diagnosis of malignant pulmonary nodules in lung cancer screening with a radiomics nomogram. Cancer Commun (Lond) 2020;40:16-24. [Crossref] [PubMed]
- Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. [Crossref] [PubMed]
- Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7:12474. [Crossref] [PubMed]
- Dercle L, Fronheiser M, Lu L, et al. Identification of non-small cell lung cancer sensitive to systemic cancer therapies using radiomics. Clin Cancer Res 2020;26:2151-62. [Crossref] [PubMed]
- Park S, Lee SM, Lee KH, et al. Deep learning-based detection system for multiclass lesions on chest radiographs: comparison with observer readings. Eur Radiol 2020;30:1359-68. [Crossref] [PubMed]
- Zhou P, Wang J, Mishail D, et al. Recent advancements in PARP inhibitors-based targeted cancer therapy. Precis Clin Med 2020;3:187-201. [Crossref] [PubMed]
- Mayerhoefer ME, Materka A, Langs G, et al. Introduction to radiomics. J Nucl Med 2020;61:488-95. [Crossref] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- Winkels M, Cohen TS. Pulmonary nodule detection in CT scans with equivariant CNNs. Med Image Anal 2019;55:15-26. [Crossref] [PubMed]
- Setio AA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 2016;35:1160-9. [Crossref] [PubMed]
- Hu J, Lu J, Tan YP, et al. Deep transfer metric learning. IEEE Trans Image Process 2016;25:5576-88. [Crossref] [PubMed]
- Xie Y, Zhang J, Xia Y. Semi-supervised adversarial model for benign-malignant lung nodule classification on chest CT. Med Image Anal 2019;57:237-48. [Crossref] [PubMed]
- Xie Y, Xia Y, Zhang J, et al. Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging 2019;38:991-1004. [Crossref] [PubMed]
- Xu X, Wang C, Guo J, et al. MSCS-DeepLN: Evaluating lung nodule malignancy using multi-scale cost-sensitive neural networks. Med Image Anal 2020;65:101772. [Crossref] [PubMed]
- Zhang N, Liang R, Gensheimer MF, et al. Early response evaluation using primary tumor and nodal imaging features to predict progression-free survival of locally advanced non-small cell lung cancer. Theranostics 2020;10:11707-18. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med 2020;382:503-13. [Crossref] [PubMed]
- Armato SG 3rd. Deep learning demonstrates potential for lung cancer detection in chest radiography. Radiology 2020;297:697-8. [Crossref] [PubMed]
- Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin 2019;69:127-57. [Crossref] [PubMed]
- Oudkerk M, Liu S, Heuvelmans MA, et al. Lung cancer LDCT screening and mortality reduction - evidence, pitfalls and future perspectives. Nat Rev Clin Oncol 2021;18:135-51. [Crossref] [PubMed]
- Jacobs C, van Ginneken B. Google's lung cancer AI: a promising tool that needs further validation. Nat Rev Clin Oncol 2019;16:532-3. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Wang X, Duan H, Li X, et al. A prognostic analysis method for non-small cell lung cancer based on the computed tomography radiomics. Phys Med Biol 2020;65:045006. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Sun W, Jiang M, Dang J, et al. Effect of machine learning methods on predicting NSCLC overall survival time based on Radiomics analysis. Radiat Oncol 2018;13:197. [Crossref] [PubMed]
- Lee J, Cui Y, Sun X, et al. Prognostic value and molecular correlates of a CT image-based quantitative pleural contact index in early stage NSCLC. Eur Radiol 2018;28:736-46. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, et al. Radiomics signature: A potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med 2018;15:e1002711. [Crossref] [PubMed]
- Xu Y, Hosny A, Zeleznik R, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res 2019;25:3266-75. [Crossref] [PubMed]
- Kim H, Goo JM, Lee KH, et al. Preoperative CT-based deep learning model for predicting disease- free survival in patients with lung adenocarcinomas. Radiology 2020;296:216-24. [Crossref] [PubMed]
- Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol 2018;51:116-28. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Mehta A, Dobersch S, Romero-Olmedo AJ, et al. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev 2015;34:229-41. [Crossref] [PubMed]
- Linehan WM, Ricketts CJ. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol 2019;16:539-52. [Crossref] [PubMed]
- Hutter C, Zenklusen JC. The Cancer Genome Atlas: creating lasting value beyond its data. Cell 2018;173:283-5. [Crossref] [PubMed]
- Yu J, Hu Y, Xu Y, et al. LUADpp: an effective prediction model on prognosis of lung adenocarcinomas based on somatic mutational features. BMC Cancer 2019;19:263. [Crossref] [PubMed]
- Li Y, Ge D, Gu J, et al. A large cohort study identifying a novel prognosis prediction model for lung adenocarcinoma through machine learning strategies. BMC Cancer 2019;19:886. [Crossref] [PubMed]
- Lee B, Chun SH, Hong JH, et al. DeepBTS: prediction of recurrence-free survival of non-small cell lung cancer using a time-binned deep neural network. Sci Rep 2020;10:1952. [Crossref] [PubMed]
- Lai YH, Chen WN, Hsu TC, et al. Overall survival prediction of non-small cell lung cancer by integrating microarray and clinical data with deep learning. Sci Rep 2020;10:4679. [Crossref] [PubMed]
- Al-Anni R, Hou J, Azzawi H, et al. Risk classification for NSCLC survival using microarray and clinical data. International Journal of Advances in Electronics and Computer Science 2019;6:20-5.
- Schalper KA, Brown J, Carvajal-Hausdorf D, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 2015;107:dju435. [Crossref] [PubMed]
- Brambilla E, Le Teuff G, Marguet S, et al. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol 2016;34:1223-30. [Crossref] [PubMed]
- Corredor G, Wang X, Zhou Y, et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res 2019;25:1526-34. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Liu M, Guo F. Recent updates on cancer immunotherapy. Precis Clin Med 2018;1:65-74. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol 2016;2:1636-42. [Crossref] [PubMed]
- Chen B, Yang L, Zhang R, et al. Radiomics: an overview in lung cancer management-a narrative review. Ann Transl Med 2020;8:1191. [Crossref] [PubMed]
- Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. [Crossref] [PubMed]
- Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol 2019;30:998-1004. [Crossref] [PubMed]
- Khorrami M, Prasanna P, Gupta A, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol Res 2020;8:108-19. [Crossref] [PubMed]
- Tian P, He B, Mu W, et al. Assessing PD-L1 expression in non-small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics 2021;11:2098-107. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Bedard PL, Hyman DM, Davids MS, et al. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020;395:1078-88. [Crossref] [PubMed]
- Song Z, Liu T, Shi L, et al. The deep learning model combining CT image and clinicopathological information for predicting ALK fusion status and response to ALK-TKI therapy in non-small cell lung cancer patients. Eur J Nucl Med Mol Imaging 2021;48:361-71. [Crossref] [PubMed]
- Song J, Wang L, Ng NN, et al. Development and validation of a machine learning model to explore tyrosine kinase inhibitor response in patients with stage IV EGFR variant-positive non-small cell lung cancer. JAMA Netw Open 2020;3:e2030442. [Crossref] [PubMed]
- Song J, Shi J, Dong D, et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin Cancer Res 2018;24:3583-92. [Crossref] [PubMed]
- Mu W, Jiang L, Zhang J, et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun 2020;11:5228. [Crossref] [PubMed]
- Wang S, Shi J, Ye Z, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 2019;53:1800986. [Crossref] [PubMed]
- Wiesweg M, Mairinger F, Reis H, et al. Machine learning-based predictors for immune checkpoint inhibitor therapy of non-small-cell lung cancer. Ann Oncol 2019;30:655-7. [Crossref] [PubMed]


