
Application of machine learning approaches to predict the 5-year survival status of patients with esophageal cancer
Introduction
Esophageal cancer (EC) is the seventh most common cancer worldwide and the sixth leading cause of cancer-related deaths. There were 544,076 new EC-related deaths in 2020, accounting for 5.5% of all new cancer-related deaths. The main pathological type of EC is squamous cell carcinoma. The incidence of esophageal squamous cell carcinoma is generally declining in some high-risk countries in Asia, but the incidence of esophageal adenocarcinoma is rising rapidly in high-income countries. Studies have shown that esophageal adenocarcinoma will surpass the incidence of esophageal squamous cell carcinoma in high-income countries in the future and become the major type of EC (1). Due to the heterogeneity of patients with EC in terms of age, pathological types, pathological stages, and treatment regimens, its prognosis varies greatly, and has received much attention.
With the development of computer technology, the application of artificial intelligence in the medical field is increasingly extensive. Machine learning (ML) is one of the best-known technologies in the field of artificial intelligence and has become a hot spot of medical research. A variety of ML techniques have been shown to be effective in predicting tumor susceptibility, recurrence, and survival of malignant tumors. In an earlier study, gradient boosting machines, support vector machines (SVM), and a custom ensemble were used to predict the survival of lung cancer patients (2). Some researchers used logistic regression (LR), artificial neural networks (ANN), and decision trees (DT) to study the survival rate of breast cancer. The results showed that compared with the two other models, DT had higher accuracy (3). In the field of EC, research of artificial intelligence has also been conducted. For example, ANN was used to predict the prognosis of EC (4).
Despite the recent popularity of deep learning neural networks, the gradient boosting methods are still recognized as the best-in-class in the field of ML when it comes to small-to-medium structured/tabular datasets, due to the lower requirement of training time and lower complexity of hyperparameter tuning. The three newly proposed gradient boosting models (GBM) of XGBoost, CatBoost, and LightGBM all excel in both speed and accuracy, and have been widely used in the medical field in recent years. In previous studies, these models have been shown to be of great value in medical imaging (5-8). In oncology, they are used in the diagnosis of malignant tumors (9,10), prediction of clinical burden before and after cancer surgery (11), and prediction of adverse reactions to adjuvant therapy (12), among other applications. They have also been shown to be effective in the prognostic prediction of malignant tumors (13,14).
In this study, information from the Surveillance, Epidemiology, and End Results (SEER) Program was used to select relevant features of patients diagnosed with EC. We used XGBoost, CatBoost, LightGBM, gradient boosting decision trees (GBDT), ANN, random forest (RF), naive Bayes (NB), and SVM to predict the 5-year survival status of patients, and the performances of these models were reported in terms of the area under the receiver operating characteristic (ROC) curve (AUC), accuracy, and logistic loss.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-1107).
Methods
Data processing
We retrieved the information of patients diagnosed with EC between 2010 and 2015 from the SEER Program, including 24 features displayed in Table 1. As the information in the SEER database does not require explicit consent from the patients, our study was not subject to the ethical approval requirements of the institutional review board.
Table 1
| Features | Number of categories | Top categorya | Frequencyb | P valuec |
|---|---|---|---|---|
| Categorical features | ||||
| Race recode (W, B, AI, API) | 4 | White | 8,941 | 0.156 |
| Sex | 2 | Male | 8,441 | 0.232 |
| Primary site-labelled | 7 | C15.5-lower third of esophagus | 6,941 | 0.841 |
| Diagnostic confirmation | 4 | Positive histology | 10,508 | 0.857 |
| ICD-O-3 Hist/behav | 47 | 8140/3: adenocarcinoma, NOS | 5,955 | <0.001 |
| Derived AJCC stage group, 7th ed (2010–2015) | 11 | IV | 3,171 | <0.001 |
| Derived AJCC T, 7th ed (2010–2015) | 11 | T3 | 4,270 | <0.001 |
| Derived AJCC N, 7th ed (2010–2015) | 4 | N1 | 4,729 | <0.001 |
| Derived AJCC M, 7th ed (2010–2015) | 2 | M0 | 7,417 | <0.001 |
| RX Summ—Surg Prim Site (1998+) | 4 | None | 7,364 | <0.001 |
| RX Summ—Scope Reg LN Sur (2003+) | 8 | None | 7,398 | <0.001 |
| RX Summ—Surg Oth Reg/Dis (2003+) | 6 | None; diagnosed at autopsy | 10,265 | 0.749 |
| SEER combined mets at DX-bone (2010+) | 2 | No | 9,832 | <0.001 |
| SEER combined mets at DX-brain (2010+) | 2 | No | 10,420 | <0.001 |
| SEER combined mets at DX-liver (2010+) | 2 | No | 9,170 | <0.001 |
| SEER combined mets at DX-lung (2010+) | 2 | No | 9,643 | <0.001 |
| CS tumor size (2004–2015) | 170 | 50 | 1,193 | <0.001 |
| CS lymph nodes (2004–2015) | 19 | 0 | 4,132 | <0.001 |
| CS mets at DX (2004–2015) | 6 | 0 | 7,417 | <0.001 |
| Sequence number | 8 | One primary only | 7,737 | <0.001 |
| Reason no cancer-directed surgery | 7 | Not recommended | 6,215 | <0.001 |
| 5-year survival | 2 | Dead | 9,048 | |
| Numerical features | ||||
| Age recode with single ages and 85+ | 66.71d | 10.89e | [18, 85]f | <0.001 |
| Regional nodes examined (1988+) | 8.66d | 21.4e | [0, 98]f | <0.001 |
| Regional nodes positive (1988+) | 70.87d | 43.38e | [0, 98]f | <0.001 |
a, the category with the highest frequency; b, the corresponding frequency; c, χ2 test; d, these data represent the mean; e, these data represent the Std.; f, these data represent the [range]. SEER, Surveillance, Epidemiology, and End Results.
The target classification of 5-year survival status for each participant (case) was calculated on the basis of survival months and vital status recode. The cases with survival months greater than or equal to 60 months were labelled as “Alive”, while the cases with “Dead” vital status recode and survival months less than 5 years were labelled as “Dead”. The cases with “Alive” vital status recode and survival months less than 5 years were removed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
Data were analyzed using the Python packages NumPy (https://numpy.org/), pandas (https://pandas.pydata.org/), and scikit-learn (https://scikit-learn.org/stable/). Categorical features were described as the number of categories, the category with the highest frequency, and the corresponding frequency. Continuous features were represented by means, standard deviations, and ranges. We performed χ2 tests between each feature and the target and 5-year survival to identify the features that are the most likely to be independent and therefore irrelevant for classification. A P value greater than 0.05 was considered a statistically significant difference.
Model building
Gradient boosting is a family of ensemble tree-based frameworks that consist of iteratively converting various weak classifiers with respect to distribution of a single final strong classifier. According to the empirical risk minimization principle, the method applies a steepest descent iteration to minimize the average value of the loss function on the training set, namely, minimize the empirical risks. The XGBoost, CatBoost, and LightGBM libraries are state-of-the-art GBMs, recognized in a number of ML and data mining challenges.
Extreme gradient boosting, or XGBoost, a scalable ML system for tree boosting, is characterized by a highly scalable end-to-end tree boosting system, theoretically justified weighted quantile sketch, the sparsity-aware algorithm, and the effective cache-aware block structure for out-of-core tree learning strategy (15).
Categorical boosting, CatBoost, is specifically for handling categorical features without preprocessing, where a new schema is used to calculate leaf values when selecting the tree structure, effectively reducing overfitting (16).
The LightGBM is a new GBM based on two novel techniques, gradient-based one-side sampling (GOSS) and exclusive feature bundling (EFB) to speed up the training process of boosting by excluding a significant proportion of data instances with small gradients and bundling mutually exclusive features to reduce the number of features, respectively (17).
In this study, the 3 models were used along with 2 other commonly used tree-based models, GBDT and RF, and 3 classical ML models, ANN, NB, and SVM, for comparison. The models were built using the Python packages XGBoost (https://xgboost.readthedocs.io/), CatBoost (https://catboost.ai/), LightGBM (https://lightgbm.readthedocs.io/), and scikit-learn. Particularly, the ANN model used in this paper was chosen from 14 different ANN structures (n-2-1, n-3-1, n-4-1, n-5-1, n-6-1, n-2-2-1, n-2-4-1, n-2-6-1, n-4-2-1, n-4-4-1, n-4-6-1, n-6-2-1, n-6-4-1, n-6-6-1; n = the number of features) as in the study of Sato et al. (4).
Notably, except for CatBoost, which is capable of handling categorical features, and LightGBM, which offers native built-in support for categorical features, all categorical features were encoded outside of the learner with the label encoding technique.
Model evaluation
Cross-validation is a form of model validation which attempts to improve on the basic methods of hold-out validation by leveraging subsets of data and an understanding of the bias/variance trade-off in order to gain a better understanding of how the models will actually perform when applied outside of the data it was trained on (18). The K-fold cross-validation is one of the most common resampling techniques used in evaluating ML models. The original sample is randomly divided into k equal sized subsamples. Among the k subsamples, a single subsample is held as the validation data to test the model, and the residual k−1 subsamples are used as training data. The cross-validation process is repeated k times, with each of the k subsamples used exactly once as the test data. The k results can then be averaged to produce a single estimation, namely, the performance measure of the model. In this paper, for the dataset with unequal class proportions, the stratified k-fold was used, where the folds were made by preserving the percentage of samples for each class. The stratified k-fold cross-validation was conducted by the Python package scikit-learn.
Hyperparameter tuning
Hyperparameters are adjustable parameters that control the model training process and dramatically influence the performance of the models. Hyperparameter tuning is an optimization problem where the objective function of optimization is unknown or a black-box function (19). The two traditional ways of performing hyperparameter optimization are grid search and random search. Grid search performs an exhaustive search through a manually specified subset of the hyperparameter space, which is computationally very expensive. In random search, the hyperparameters are randomly selected, and not every combination of parameters is tried. These two methods do not learn from previous results. Conversely, Bayesian optimization iteratively evaluates a promising hyperparameter configuration based on prior information, including previous hyperparameter configurations and the corresponding objective function loss of the model, and then updates it. Bayesian optimization allows exploration (trying new hyperparameter values) and exploitation (using hyperparameter configuration resulting in the lowest objective function loss) to be naturally balanced during the search. In practice, it is shown that compared to grid search and random search, Bayesian optimization is able to obtain better results in fewer evaluations, due to the ability to reason about the quality of trials before they are run.
In this paper, Bayesian optimization was implemented by a hyperparameter optimization framework Optuna (20), using the Python package optuna 2.8.0 (https://optuna.org/), where we can define the parameter space and the trials, adopt state-of-the-art algorithms for sampling hyperparameters, and efficiently prune unpromising trials. A trial is a single execution of the objective function, which was defined as the average logistic loss of the 5-fold cross-validation of the model in this paper. In each trial, the hyperparameters were selected from the parameter space according to the prior information, and then the stratified 5-fold cross-validation was executed to produce the average logistic loss to estimate the model with the selected hyperparameters. The parameter spaces of each model are shown in Table 2 and the number of trials was set at 100. After 100 trials, the hyperparameters with minimum average logistic loss were chosen for the final model comparison. Notably, there were fewer hyperparameters used for SVM and NB, obtained by the trial-and-error method.
Table 2
| Classifier | Training parameters | Searching space | Best parameters |
|---|---|---|---|
| XGBoost | n_estimators | [100, 10,000] | 1,169 |
| learning_rate | [0.001, 0.5] | 0.1 | |
| max_depth | [1, 10] | 5 | |
| subsample | [0.25, 0.75] | 0.62 | |
| colsample_bytree | [0.05, 0.5] | 0.49 | |
| colsample_bylevel | [0.05, 0.5] | 0.41 | |
| CatBoost | n_estimators | [100, 10,000] | 1,642 |
| learning_rate | [0.001, 0.5] | 0.1 | |
| max_depth | [0, 5] | 3 | |
| reg_lambda | [1e−8, 10] | 0.3455 | |
| LightGBM | n_estimators | [100, 10,000] | 3,248 |
| learning_rate | [0.001, 0.5] | 0.0316 | |
| max_depth | [1, 10] | 5 | |
| num_leaves | [1, 300] | 16 | |
| lambda_l1 | [1e−8, 10] | 0.52 | |
| lambda_l2 | [1e−8, 10] | 0.2 | |
| GBDT | n_estimators | [100, 5,000] | 1,340 |
| learning_rate | [0.001, 0.5] | 0.0023 | |
| max_depth | [1, 20] | 11 | |
| max_leaf_nodes | [2, 100] | 23 | |
| subsample | [0.25, 0.75] | 0.27 | |
| RF | n_estimators | [100,10,000] | 200 |
| max_depth | [1, 10] | 6 | |
| min_samples_split | [2, 11] | 2 | |
| min_samples_leaf | [1, 10] | 4 |
GBDT, gradient boosting decision trees; RF, random forest.
Feature importance
The ML models are able to build non-linear and complex relationships between the features and target. However, it is a challenge to understand why a model makes a certain prediction and access the global feature importance, which is, in a way, a black box. To address this problem, Lundberg and Lee presented a unified framework, SHapley Additive exPlanations (SHAP), to improve the interpretability (21). The SHAP value is the average marginal contribution of a feature value across all possible coalitions. The interpretation of the SHAP value for feature value is: the value of the -th feature contributed to the prediction of this particular instance compared to the average prediction for the dataset. In this paper, the SHAP values were calculated by the Python package SHAP (https://shap.readthedocs.io/).
Results
A total of 10,588 patients (cases) were selected from the SEER database, whose years of diagnoses were from 2010 to 2015. The samples consisted of two classes (9,048 cases with “Dead” status and 1,540 cases with “Alive” status), which showed the imbalance of the sample number. A set of features were selected from the dataset, consisting of 21 categorical features and 3 numerical features, as displayed in Table 1. There were 5 features with p-values greater than 0.05. We compared the performances of each model trained by the complete dataset and the dataset with the non-significant features removed.
Hyperparameter tuning
A diagram of the process of hyperparameter tuning of the LightGBM model is shown in Figure 1. Lambda_l1 and lambda_l2 are 2 hyperparameters in the LightGBM model. There are 100 dots in Figure 1 and each dot represents a trial, the location of which shows the corresponding lambda_l1 and lambda_l2 values. The shade of blue indicates the range of the objective values for the trials. In addition, it can be observed that the lighter the color, the denser the dots. Bayesian optimization balances between exploration (hyperparameter configuration for which the objective value is most uncertain) and exploitation (hyperparameter configuration expected close to the optimum). That is to say, some of the trials might concentrate on hyperparameter values around the local minimum, while others would try new hyperparameter configurations. Therefore, in the area with low objective value, the dots would assemble, and the hyperparameters near the dots with high value would not be selected to be trialed.
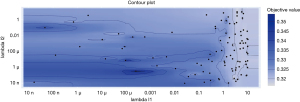
The best hyperparameters found in the hyperparameter tuning processes of XGBoost, CatBoost, LightGBM, GBDT, and RF are shown in Table 2. The descriptions of all training parameters are displayed in Table S1.
Model performance
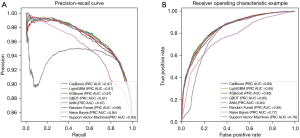
In Table 3 and Figure 2, we summarized the performance of 8 models in terms of ROC, AUC, accuracy, logistic loss, and precision-recall curve, which are the average results of 5-fold cross-validation. The 8 models could be sorted from the best to the poorest as follows: XGBoost > LightGBM > CatBoost > GBDT > ANN > RF > NB > SVM, where the three variants of GBDT performed with little difference, and the curves in both graphs (Figure 2) almost coincided. Moreover, the performances of each model trained by the complete dataset were better than those trained by the dataset with the non-significant features removed. After overall consideration of the several performance metrics, the XGBoost and the complete dataset were chosen to build the final model to predict the 5-year survival status.
Table 3
| Classifier | The complete dataset |
The dataset with the non-significant features removed |
||||||
|---|---|---|---|---|---|---|---|---|
| AUC | Accuracy | Logistic loss | AUC | Accuracy | Logistic loss | |||
| XGBoost | 0.852 | 0.875 | 0.301 | 0.845 | 0.871 | 0.307 | ||
| LightGBM | 0.850 | 0.875 | 0.302 | 0.844 | 0.870 | 0.308 | ||
| CatBoost | 0.849 | 0.874 | 0.304 | 0.843 | 0.871 | 0.308 | ||
| GBDT | 0.846 | 0.875 | 0.307 | 0.842 | 0.871 | 0.311 | ||
| ANNa | 0.844 | 0.871 | 0.308 | 0.833 | 0.869 | 0.316 | ||
| RF | 0.838 | 0.865 | 0.319 | 0.838 | 0.865 | 0.319 | ||
| NB | 0.833 | 0.769 | 1.766 | 0.833 | 0.769 | 1.766 | ||
| SVM | 0.789 | 0.855 | 0.364 | 0.789 | 0.855 | 0.363 | ||
a, the ANN structure with the best AUC is n-4-4-1. AUC, area under the receiver operating characteristic curve; GBDT, gradient boosting decision trees; RF, random forest; NB, naive Bayes; ANN, artificial neural networks; SVM, support vector machines.
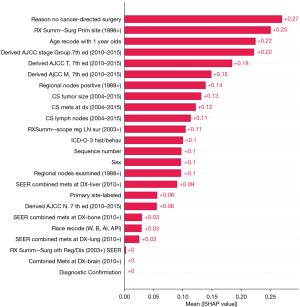
Feature importance
SHAP values represent a feature’s responsibility for a change in the model output. Therefore, as depicted in Figure 3, the features with larger mean SHAP value were more important in model prediction, where “Reason no cancer-directed surgery” was the most important.
Because 5-year survival status was processed by label encoding, where “Alive” was labelled as “0” and “Dead” as “1”, the positive SHAP value increased the probability of death, as the higher the value, the higher the risk, and vice versa. Therefore, from Figure 4, high values of “Reason no cancer-directed surgery” and “RX Summ—Surg Prim Site (1998+)” increased the probability of “Alive”. Specifically, for the categorical feature “Reason no cancer-directed surgery”, category “Not recommended” with 6,215 cases was labelled as “1”, and “Surgery performed” with 3,224 cases was labelled as “6”, indicating that “Surgery performed” increased the probability of “Alive”, and “Not recommended” was likely to lead to “Dead”. For the categorical feature “RX Summ—Surg Prim Site (1998+)”, category “None” with 7,364 cases was labelled as “0”, and category “Site-specific codes-resection” with 3,171 cases was labelled as “2”, demonstrating that “Site-specific codes-resection” increased the probability of “Alive”. Moreover, from the aspect of age, the older the age, the higher the risk of death. For “Derived AJCC stage group, 7th ed (2010–2015) T and M”, the higher the stage, the higher the risk of death.
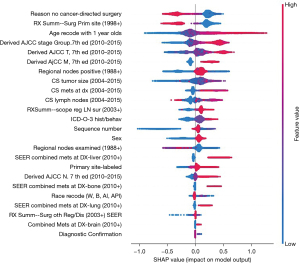
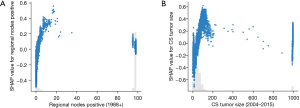
Unlike the previous features, the color distribution was irregular for “Regional nodes positive” and “CS tumor size”, implying that the values of the features were not correlated linearly with the SHAP values. To explore the reason, we plotted the SHAP values of these two features versus the values of the features for all participants in the dataset (Figure 5). Values 95–99 for “Regional nodes positive” refer to cases where no regional nodes were removed or the number of nodes was unknown/not stated. Values 991–995 for “CS tumor size” refer to cases where tumor size was described as less than 1 to 5 cm, respectively. Value 990 means microscopic focus or foci only. If no size of focus is given, values 996–998 indicate site-specific codes where needed, and value 999 indicates unknown. The figures displayed that, except for the special values mentioned above, the higher the values of the features, the higher risk of death.
Discussion
This study attempted to use ML to predict the 5-year survival status of EC patients, and successfully constructed a 5-year survival status model of EC. The model demonstrated good predictive performance through routine clinical data.
In this study, discrimination of the performance of the three newly developed variants of GBM showed that they were similar, with little variability in ROC, precision-recall curve (Figure 2), and three other metrics (Table 3). They outperformed other models, including the ANN used by Sato et al. (4). The prediction result (AUC =0.88) in the study of Sato et al. (4) was higher than that in this paper, but there were more features (199 features) used to train the model, providing more information and thus better prediction of outcomes. Besides, the precision-recall curve showed that the three variants are effective in predicting the imbalanced dataset; however, NB and SVM had poor performance in the class accuracy of the small number of samples caused by the imbalance of sample number.
Furthermore, the fact that the three newly developed variants of GBM trained by the complete dataset performed better than those trained by the dataset with non-significant features removed implied that for these three algorithms, the more comprehensive the information, the better the model will perform, and redundant information does not interfere with model prediction. Cross-validation was used to avoid overfitting, which significantly improves the classification accuracy and generalization capability. The hyperparameter tuning based on Bayesian optimization is a more efficient method compared with grid search and random search, and is a necessary process before constructing the final model, which is able to markedly improve the model performance.
Interpreting a model’s prediction outcome is very important in many application scenarios for ML. The trade-off between accuracy and interpretability of a model’s output is always a problem for researchers in many fields. Both SHAP and Local Interpretable Model-agnostic Explanations (LIME) are popular approaches for model interpretability. The LIME approach builds sparse linear models around each prediction to explain how the black box model works in that local vicinity. Lundberg and Lee showed that SHAP provides the only guarantee of accuracy and consistency, while LIME is actually a subset of SHAP but lacks the same properties (21). However, SHAP is an exhaustive method which considers all possible predictions for an instance using all possible combinations of inputs, and is therefore time consuming compared with LIME. Since the sample size of 10,588 is a small dataset in data mining fields, SHAP was used in this paper. The interpretations provided by SHAP are presented as follows.
Access to surgical treatment is an important factor in model prediction. Compared with patients with stage I–III EC who did not undergo surgery, those treated with surgery had better long-term survival (22). In recent years, with the development of neoadjuvant therapy, many patients with locally advanced EC have gained the opportunity of surgical treatment. A previous study showed that an en bloc resection at the original operation site is the most significant predictor of prolonged survival (23).
According to the model, the earlier the tumor stage lowers the predicted probability of death. On the contrary, if the tumor stage is late, it could easily lead to death. Lymph node metastasis is a key factor in evaluating the prognosis of patients with EC (24). Studies have shown that extensive lymph node dissection is associated with improved survival in patients with stage II–III esophageal squamous cell carcinoma (25). Under the premise of sufficient lymph node dissection, the ratio of the number of positive lymph nodes to the total number of lymph nodes has been used as an important indicator to predict the prognosis of EC in many studies. Notably, as part of tumor staging, N status did not contribute significantly to predicting outcomes compared to T and M. The possible reason is that there is a linear relationship between lymph node metastasis and tumor stage. The effect of lymph node metastasis on outcome prediction was reflected by tumor stage. According to the results, tumor staging played an important role in predicting 5-year survival.
Age was one of the most important factors in model prediction. The prevalence of EC is higher in elderly patients, and many studies have shown that the increase in age is associated with the decrease of 1- and 5-year survival (26,27). For patients with locally advanced EC, postoperative survival was negatively correlated with age.
The main limitations of this study are as follows. Some critical factors that are strong predictors of survival and patient outcomes are unavailable in the SEER database, such as surgical methods postoperative complications, and more importantly, radiation and chemotherapy information. Some technical advances in surgical methods such as preoperative simulation, robot-assisted thoracoscopic esophagectomy, and intraoperative real-time navigation may decrease the morbidity and mortality rate of surgery for EC and hopefully improve oncological outcomes (28). Susceptibility to a variety of complications is also one of the characteristics of EC. Anastomotic leaks, chyle leaks, cardiopulmonary complications, and later functional issues after esophagectomy may result in long-term sequelae and even death (29). The effect of radiotherapy and chemotherapy on patients with EC is still a research hotspot. For locally advanced esophagogastric junction patients, neoadjuvant chemoradiotherapy results in a better survival rate than neoadjuvant chemotherapy (30). At the same time, radiotherapy or neoadjuvant chemoradiotherapy may also increase the incidence of cardiac and pulmonary complications (31,32). These factors have a negative impact on the accuracy of the prediction. The introduction of more features for analysis will be crucial for building effective prediction models in the future. If the features mentioned above become available, the model should be trained from scratch, including the process of feature selection, hyperparameter tuning, and model evaluation.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-1107
Peer Review File: Available at https://dx.doi.org/10.21037/jtd-21-1107
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-1107). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Since any information in the SEER database does not require explicit consent from the patients, our study was not subject to the ethical approval requirements of the institutional review board. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arnold M, Laversanne M, Brown LM, et al. Predicting the future burden of esophageal cancer by histological subtype: international trends in incidence up to 2030. Am J Gastroenterol 2017;112:1247-55. [Crossref] [PubMed]
- Lynch CM, Abdollahi B, Fuqua JD, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inform 2017;108:1-8. [Crossref] [PubMed]
- Delen D, Walker G, Kadam A. Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med 2005;34:113-27. [Crossref] [PubMed]
- Sato F, Shimada Y, Selaru FM, et al. Prediction of survival in patients with esophageal carcinoma using artificial neural networks. Cancer 2005;103:1596-605. [Crossref] [PubMed]
- Roy SD, Das S, Kar D, et al. Computer aided breast cancer detection using ensembling of texture and statistical image features. Sensors (Basel) 2021;21:3628. [Crossref] [PubMed]
- Lin F, Cui EM, Lei Y, et al. CT-based machine learning model to predict the Fuhrman nuclear grade of clear cell renal cell carcinoma. Abdom Radiol (NY) 2019;44:2528-34. [Crossref] [PubMed]
- Tahmassebi A, Wengert GJ, Helbich TH, et al. impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Invest Radiol 2019;54:110-7. [Crossref] [PubMed]
- Jiang YQ, Cao SE, Cao S, et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J Cancer Res Clin Oncol 2021;147:821-33. [Crossref] [PubMed]
- Carvalho ED, Filho AOC, Silva RRV, et al. Breast cancer diagnosis from histopathological images using textural features and CBIR. Artif Intell Med 2020;105:101845 [Crossref] [PubMed]
- Zhang Y, Feng T, Wang S, et al. A novel XGBoost method to identify cancer tissue-of-origin based on copy number variations. Front Genet 2020;11:585029 [Crossref] [PubMed]
- Low CA, Li M, Vega J, et al. Digital biomarkers of symptom burden self-reported by perioperative patients undergoing pancreatic surgery: prospective longitudinal study. JMIR Cancer 2021;7:e27975 [Crossref] [PubMed]
- Ahn BC, So JW, Synn CB, et al. Clinical decision support algorithm based on machine learning to assess the clinical response to anti-programmed death-1 therapy in patients with non-small-cell lung cancer. Eur J Cancer 2021;153:179-89. [Crossref] [PubMed]
- Osman MH, Mohamed RH, Sarhan HM, et al. Machine learning model for predicting postoperative survival of patients with colorectal cancer. Cancer Res Treat 2021; Epub ahead of print. [Crossref] [PubMed]
- Xu Y, Ju L, Tong J, et al. Machine learning algorithms for predicting the recurrence of stage IV colorectal cancer after tumor resection. Sci Rep 2020;10:2519. [Crossref] [PubMed]
- Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016:785-94.
- Prokhorenkova L, Gusev G, Vorobev A, et al. CatBoost: unbiased boosting with categorical features. arXiv 2017:1706.09516.
- Ke G, Meng Q, Finley T, et al. LightGBM: a highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst 2017;30:3146-54.
- Forman BD, Eidson K, Hagan BJ. Measuring perceived stress in adolescents: a cross validation. Adolescence 1983;18:573-6. [PubMed]
- Ahn CW, Ramakrishna RS, Goldberg DE. Real-coded Bayesian optimization algorithm: bringing the strength of BOA into the continuous world. In: Genetic and Evolutionary Computation Conference. Berlin, Heidelberg: Springer, 2004:840-51.
- Akiba T, Sano S, Yanase T, et al. Optuna: A next-generation hyperparameter optimization framework. In: Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2019:2623-31.
- Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Proceedings of the 31st International Conference on Neural Information Processing Systems, 2017:4768-77.
- Schlottmann F, Gaber C, Strassle PD, et al. Disparities in esophageal cancer: less treatment, less surgical resection, and poorer survival in disadvantaged patients. Dis Esophagus 2020;33:doz045 [Crossref] [PubMed]
- Ghaly G, Kamel M, Nasar A, et al. Locally advanced esophageal cancer: What becomes of 5-year survivors? J Thorac Cardiovasc Surg 2016;151:726-32. [Crossref] [PubMed]
- Mariette C, Piessen G, Briez N, et al. The number of metastatic lymph nodes and the ratio between metastatic and examined lymph nodes are independent prognostic factors in esophageal cancer regardless of neoadjuvant chemoradiation or lymphadenectomy extent. Ann Surg 2008;247:365-71. [Crossref] [PubMed]
- Ho HJ, Chen HS, Hung WH, et al. Survival impact of total resected lymph nodes in esophageal cancer patients with and without neoadjuvant chemoradiation. Ann Surg Oncol 2018;25:3820-32. [Crossref] [PubMed]
- Chen MF, Yang YH, Lai CH, et al. Outcome of patients with esophageal cancer: a nationwide analysis. Ann Surg Oncol 2013;20:3023-30. [Crossref] [PubMed]
- Qiu MJ, Yang SL, Wang MM, et al. Prognostic evaluation of esophageal cancer patients with stages I-III. Aging (Albany NY) 2020;12:14736-53. [Crossref] [PubMed]
- Beukema JC, van Luijk P, Widder J, et al. Is cardiac toxicity a relevant issue in the radiation treatment of esophageal cancer? Radiother Oncol 2015;114:85-90. [Crossref] [PubMed]
- Mboumi IW, Reddy S, Lidor AO. Complications after esophagectomy. Surg Clin North Am 2019;99:501-10. [Crossref] [PubMed]
- Thomas M, Defraene G, Lambrecht M, et al. NTCP model for postoperative complications and one-year mortality after trimodality treatment in oesophageal cancer. Radiother Oncol 2019;141:33-40. [Crossref] [PubMed]
- Kikuchi H, Takeuchi H. Future perspectives of surgery for esophageal cancer. Ann Thorac Cardiovasc Surg 2018;24:219-22. [Crossref] [PubMed]
- Li J, Zhao Q, Ge X, et al. Neoadjuvant chemoradiotherapy improves survival in locally advanced adenocarcinoma of esophagogastric junction compared with neoadjuvant chemotherapy: a propensity score matching analysis. BMC Surg 2021;21:137. [Crossref] [PubMed]
(English Language Editors: C. Betlazar-Maseh and J. Jones)


