
Clinical impact of nosocomial infection with pandemic influenza A (H1N1) 2009 in a respiratory ward in Guangzhou
Introduction
When novel respiratory-transmitted viruses emerge, such as the pandemic (H1N1) 2009 influenza A virus [A(H1N1)pdm09] in 2009 and notorious severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) in 2019, the population is generally susceptible because of a lack of immunity to novel virus strains (1-3). In addition to infecting individuals with underlying conditions, elderly individuals, and pregnant women, A(H1N1)pdm09 also infects young healthy individuals without underlying diseases. The A(H1N1)pdm09 strain is characterized by high transmissibility, particularly among school-aged children and household members (4-6).
Influenza viruses are spread via exposure to large-particle respiratory droplets, direct contact with patients with influenza, or indirect contact with contaminated surfaces or environments. All routes of influenza virus transmission are universally present in hospitals. Close contact between health care workers and inpatients facilitates the transmission of influenza viruses. In areas with inadequate medical resources, crowded rooms and closed environments also contribute to the long-term persistence of influenza viruses (7). Hence, it is common for influenza outbreaks to occur in health care settings. The rates of infection among patients can reach 50% in wards and 20% throughout the hospital during influenza outbreaks in the community (8).
Sporadic nosocomial influenza infections have been reported in mainland China, including in Xi’an and Taizhou (9-11). Yang et al. found that lymphocytopenia, hypoalbuminemia and pleural effusion were independent factors that can help identify patients at high risk of hospital-acquired influenza A infection, which can prolong the hospital stay and is associated with high mortality (9). However, reports on nosocomial influenza outbreaks in southern Chinese hospitals are limited. A better understanding of the clinical impact, infection control measures, and the epidemiological and molecular characteristics of the virus may help clinicians prevent and control nosocomial influenza infections. During a period in which surveillance of influenza among hospitalized patients was being performed, an outbreak of nosocomial influenza occurred. We conducted an observational study of this nosocomial influenza outbreak to evaluate the effects of the infection control measures taken and the clinical impacts in patients in a respiratory department of a tertiary hospital in Guangzhou, southern China.
We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-897).
Methods
Study population
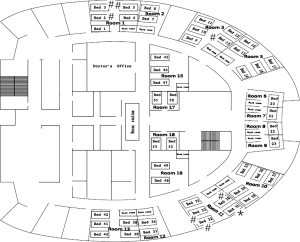
Between January and February 2016, a nosocomial influenza outbreak occurred in the respiratory department of a 1,520-bed hospital in Guangzhou, China. The respiratory department has 16 rooms and 47 beds. The distribution of rooms is shown in Figure 1. Patients who developed influenza-like illness (ILI) more than 48 hours after hospitalization were enrolled as patients with suspected cases and were followed until discharge. ILI was defined by the presence of a fever (≥38 °C), at least one systemic symptom (chills, headache, myalgia or fatigue) and at least one respiratory symptom (cough, sore throat or coryza). Nasopharyngeal swab samples were tested for the presence of influenza A/B viruses with a rapid influenza antigen test (Clearview, UK), real-time reverse transcription polymerase chain reaction (RT-PCR) and viral culture. ILI cases were confirmed if one of the abovementioned tests was positive; otherwise, the cases were defined as probable. Patients in the index ward were defined as group A, while those in other wards were defined as group B to reduce bias in the investigation of possible transmission routes of influenza. Patients in group A were monitored for the shedding of influenza virus in throat swabs by RT-PCR. Patients who were unwilling to provide informed consent were excluded from the study.
Epidemiological and clinical data (including demographic characteristics, underlying diseases, influenza vaccination status, duration of hospitalization, contact with patients with ILIs, onset and clinical course of ILI, excess cost due to ILI, delay in discharge from hospital, etc.) of all patients with suspected cases were recorded by a trained physician to evaluate the clinical impact. The cost for each patient was defined as medical fees during hospitalization, such as those for medical care, laboratory testing, inspections, drugs, etc. Living expenses were not included.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study was approved by the Ethics Committee of Scientific Research Projects of the First Affiliated Hospital of Guangzhou Medical University (No. 2014.14) and informed consent was obtained from all the patients.
RT-PCR
All swab samples were tested for the presence of influenza A virus, influenza B virus and A(H1N1)pdm09 with the Flu A/Flu B/H1 kit and an ABL7500 system in accordance with the manufacturer’s protocols (Guangzhou Institute of Respiratory Disease Medical Technology, Co., Ltd., Guangzhou, China), as described elsewhere (12).
Viral sequencing and phylogenetic analysis
To verify that the nosocomial cases of influenza originated from the same index patient, we further sequenced the viral strains and performed phylogenetic analysis. Influenza viruses were isolated from all samples using Madin-Darby canine kidney (MDCK) cells. Viral cultures from positive samples were collected and stored at −70 °C prior to molecular sequencing.
Viral RNA was extracted from the viral culture medium using a QIAamp Viral RNA Mini Kit (Qiagen Co., Ltd., Germany) in accordance with the manufacturer’s protocols. A QIAGEN OneStep RT-PCR Kit was used to amplify the target genes. The primers for the hemagglutinin (HA) and neuraminidase (NA) genes were designed according to the recommendations of the National Institutes of Health, and the HA sequence was divided into HA1 and HA2 sequences (Table 1). The drug resistance mutation H275Y was covered by the NA primers. The conditions for amplification were as follows: 50 °C for 15 min; 94 °C for 2 min; 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; and 72 °C for 10 min for HA1 and HA2 and 50 °C for 15 min; 94 °C for 2 min; 35 cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 90 s; and 72 °C for 10 min for NA. The PCR products were electrophoresed in a 1.2% agarose gel for quality control and then purified with an agarose gel recovery kit.
Table 1
| Name | Primer sequence | Length of target fragment (bp) |
|---|---|---|
| HA1-F | 5'-ATACGACTAGCAAAAGCAGGGG-3' | 1,162 |
| HA1-R | 5'-TGCTCATTTTGATGGTGATAACCG-3' | |
| HA2-F | 5'-ATCCGATCACAATTGGAAAATGTCC-3' | 812 |
| HA2-R | 5'-GTGTCAGTAGAAACAAGGGTGTTT-3' | |
| NA-F | 5'-AGCAAAAGCAGGAGT-3' | 1493 |
| NA-R | 5'-AGTAGAAACAGGAG-3' |
HA, hemagglutinin; NA, neuraminidase; A(H1N1)pdm09, the pandemic (H1N1) 2009 influenza A virus; F, forward; R, reverse.
The PCR products were sequenced on an ABI 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, USA). The sequences were assembled, edited and aligned with BioEdit, and a phylogenetic tree was constructed with Molecular Evolutionary Genetics Analysis version 5.05 (MEGA 5.05) software. The reliability of all the phylogenetic groupings was determined through a bootstrap resampling analysis (1,000 replicates). Viral strains were classified according to the protocol for influenza A virus global swine H1 clade classification (Influenza Research Database).
Statistical analyses
This was a prospective study of nosocomial influenza in which the goal was to enroll all influenza cases and evaluate the clinical impacts rather than evaluate the efficacy of infection control measures. Therefore, no sample size calculation was performed. Quantitative data are presented as medians and interquartile ranges (IQRs) or means ± standard deviations (SDs), and categorical variables are reported as frequencies and percentages.
Results
Clinical characteristics
The suspected index patient (patient 1) was a female patient with tuberculous stenosis of the left main bronchus who developed high-grade fever (the clinical presentation was consistent with an ILI) on 21 January 2016. Influenza was suspected, which prompted nasopharyngeal swab sampling. Oseltamivir was administered despite a negative result on the rapid influenza antigen test. Three days later, three of four other patients in the same room (Room 11) successively developed ILIs (patients 2, 4 and 6), and one of the four patients had a positive rapid influenza antigen test result. These findings prompted the suspicion of a nosocomial influenza outbreak. Additionally, patients 3 and 5 in Room 1 also developed symptoms of ILI after patient 3 underwent bronchoscopy. Ultimately, eight hospitalized patients in three rooms developed ILIs during the outbreak and were included in our analysis (Figure 1). Seven of the eight patients underwent nasopharyngeal swab sampling and tested positive for A(H1N1)pdm09 according to real-time RT-PCR (Table 2). Therefore, an infection control measures bundle was immediately implemented to minimize further nosocomial spread of the pathogen; the bundle included isolating the patients with ILIs, performing rapid influenza antigen testing, initiating anti-influenza treatment, enforcing hand hygiene, monitoring the temperature of medical staff, and enforcing stringent visitor control. Patients in the same room could communicate with each other before the influenza outbreak, but it was suggested that communication be avoided after the influenza outbreak, especially for fever patients. During the outbreak, Rooms 12 and 15 were used as isolation rooms, and five patients were transferred to these rooms once the clinical diagnosis was confirmed (Figure 2).
Table 2
| Patient ID | Bed and room No. | Sex | Age (years) | Bed transfer date | Bed and room transfer No. | Comorbidity | Vaccination for influenza | Date of onset | Fever (°C) | LOS before symptom onset (days) | Contact with patients with ILIs | Rapid antigen test | Culture | Real-time RT-PCR | Antiviral therapy | Case classification | Delay in discharge from hospital (days) | Excess cost due to influenza [Yuan (RMB)] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Bed 31 in room 11 | F | 38 | 26-Jan | Bed 37 in room 12 | Bronchogenic tuberculosis | No | 21-Jan | 39 | 35 | No | − | − | + | Yes | Confirmed | 7 | 2,440 |
| 2 | Bed 32 in room 11 | M | 45 | 30-Jan | Bed 47 in room 15 | Penicilliosis marneffei infection | No | 24-Jan | 39 | 9 | Yes | − | − | + | Yes | Confirmed | 11 | 11,182 |
| 3 | Bed 5 in room 1 | M | 30 | NA | NA | Pulmonary cryptococcosis | No | 24-Jan | 38.4 | 5 | No | − | NT | NT | No | Probable | 3 | 527 |
| 4 | Bed 30 in room 11 | F | 60 | 26-Jan | Bed 36 in room 12 | Tracheostenosis (after endotracheal intubation) | No | 25-Jan | 39 | 4 | Yes | + | + | + | Yes | Confirmed | 7 | 4,532 |
| 5 | Bed 2 in room 1 | M | 40 | 30-Jan | Bed 45 in room 15 | Bronchiectasis | No | 25-Jan | 39.3 | 5 | Yes | − | + | + | Yes | Confirmed | 8 | 4,275 |
| 6 | Bed 33 in room 11 | M | 41 | NA | NA | Pulmonary cryptococcosis | No | 25-Jan | 39.4 | 4 | Yes | NT | + | + | Yes | Confirmed | 4 | 1,304 |
| 7 | Bed 13 in room 3 | F | 55 | 2-Feb | Bed 46 in room 15 | Bronchiectasis | No | 27-Jan | 39.4 | 8 | No | − | + | + | Yes | Confirmed | 8 | 9,784 |
| 8 | Bed 3 in room 1 | M | 36 | NA | NA | Interstitial pneumonia | No | 1-Feb | 38.5 | 8 | No | − | − | + | Yes | Confirmed | 2 | 1,908 |
−, negative; +, positive. ILI, influenza-like illness; LOS, length of stay; NT, not test. M, male; F, female.
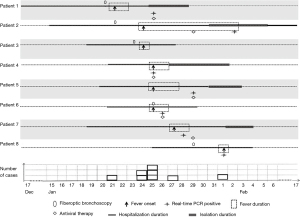
All patients developed fever, fatigue, and headache. Most patients had systemic or respiratory symptoms (e.g., cough, myalgia). Six patients had available serum procalcitonin (PCT) and C-reactive protein (CRP) levels, and their PCT levels were normal or slightly elevated, while their CRP levels were markedly elevated (l45.4±33.1 ng/mL).
Because five of eight patients had undergone fiberoptic bronchoscopy within 3 days before developing fever (Figure 2), we assessed whether bronchoscopy contributed to the transmission of influenza. A survey was conducted to investigate patients who had undergone bronchoscopy. During this period, bronchoscopy was performed by five specialized doctors. The management of bronchoscopes followed a standard operation procedure. Fifty-four patients (group B) had undergone bronchoscopy, of whom 6 (11.1%) developed transient fever. These patients were distributed in various wards of the hospital. None of the other patients who were hospitalized in the same ward as the 6 patients in group B developed fever. Only 1 patient (1.9%) experienced worsening respiratory symptoms. Only 3 patients (5.6%) received a short-term course of empirical antibiotics, and all 6 patients (11.1%) recovered from fever.
All laboratory-confirmed cases in patients in group A were treated with oseltamivir for a mean of 4 days (2 to 6 days). All patients recovered from nosocomial influenza infection and were discharged home. Patient 2 had a prolonged period of viral shedding. He had bronchiectasis and Penicillium marneffei infection, which are common in immunocompromised patients. His fever was relieved at 7 days after onset. However, the viral test was still positive 11 days after the onset of fever. The duration of viral shedding was significantly longer than that of fever. Compared to the expected duration of hospitalization in patients without fever, the duration of hospitalization in patient with fever was prolonged by a median of 7.0 (3.75–8) days, and the mean excess cost was 3,358±1,399 RMB.
During the outbreak, none of the health care workers reported symptoms of ILIs; therefore, nasopharyngeal swab sampling and viral detection were not performed. Oseltamivir was not prescribed for patients other than those with ILIs in the index ward or health care workers.
Phylogenetic analysis
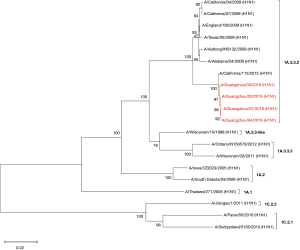
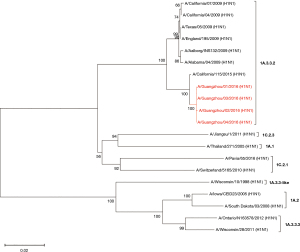
Influenza virus isolates were successfully recovered from four of seven patients who tested positive for A(H1N1)pdm09. The HA and NA genes of these four strains were sequenced. The phylogenetic analysis of the concatenated HA and NA sequences revealed that these strains were clustered in the same clade and were genetically similar to the A/California/115/2015 strain, which is classified in the 1A.3.3.2 lineage (Figures 3,4) and negative for the drug resistance mutation H275Y.
Discussion
Here, we report a case series of hospitalized patients with nosocomial A(H1N1)pdm09 infections in a ward of the respiratory department of a tertiary hospital in Guangzhou city. By adopting an infection control measure bundle, the nosocomial influenza outbreak was effectively contained. None of the patients had concomitant disorders, and all the patients recovered from influenza after antiviral treatment. Moreover, all but one patient tested positive for A(H1N1)pdm09, and most strains were confirmed to belong to an identical cluster based on the molecular phylogenetic analysis of four influenza virus isolates.
Nosocomial influenza is common because of the transmissibility of the influenza virus and susceptibility of the hospitalized population. Shedding of influenza virus particles occurs in asymptomatic patients during the incubation period (13). These patients are unlikely to be isolated before diagnosis. Hence, these patients are important sources of nosocomial influenza virus infections. Most patients in respiratory wards have underlying chronic pulmonary diseases and are more susceptible to developing severe influenza than healthy subjects. Moreover, hospitalized patients are likely to reside in crowded rooms where ventilation is poor, which facilitates the transmission of the influenza virus between the index patient and susceptible individuals. In fact, the influenza virus can be easily transmitted through large-particle respiratory droplets produced by the index patient when they cough or sneeze. Indirect contact transmission can occur when influenza viruses on a contaminated surface or substance are transferred to the mucous membranes in the nose, mouth or eyes. In this nosocomial influenza outbreak, some patients who developed ILIs were hospitalized in the same room as the index patient. Therefore, these patients with secondary cases likely came into contact with a contaminated environment or inhaled air contaminated with respiratory droplets containing the influenza virus.
The virus isolates were classified into the same lineage by phylogenetic analysis, suggesting that they originated from the same ancestor. However, how the influenza virus was transmitted from the patients in Room 11 to patient 3 in Room 1 and patient 7 in Room 3 remains unclear. Early in the outbreak, it was thought that these patients were infected during bronchoscopy. However, there were no secondary cases associated with the 6 primary fever patients in group B in the other wards. Hence, transmission might have occurred through respiratory droplet exposure or indirect contact with contaminated surfaces. Health care workers might have also contributed to the transmission of the influenza virus through indirect contact. A similar phenomenon was reported in a study by Eibach, in which three H3N2 outbreaks occurred in an acute-care geriatric department. They found that two outbreaks lasted for only 2 to 3 days because all the patients were promptly isolated, whereas the third lasted for at least 10 days because only one patient was isolated (14). This was different from the nosocomial transmission of Middle East respiratory syndrome coronavirus, for which the risk factors for transmission include the location in which exposure occurs (15). Hence, the early diagnosis of influenza and quarantining of affected patients are crucial to minimizing the likelihood of viral transmission. The prompt quarantining of affected patients results in rapid control of outbreaks within respiratory wards. This infection control method was also successful for the early control of nosocomial SARS-CoV-2 transmission, which occurred among patients via respiratory exposure or indirect contact with an index patient (16,17).
Generally, influenza virus shedding occurs from the day before symptom onset until 5 days after symptom onset (18). It is recommended that hospitalized influenza patients, whether they have suspected or confirmed cases, are isolated considering standard isolation precautions and precautions targeting control of the dissemination of respiratory droplets for 7 days after the onset of symptoms (19). However, the duration of viral shedding is prolonged in immunocompromised patients and critically ill patients (20-22), as was observed in patient 2 in our study. As recommended by the Centers for Disease Control and Prevention in the US (23), patients with influenza can be discharged when their respiratory symptoms and fever clinically improve or at 7 days after symptom onset. However, when patients continue to exhibit symptoms or virus shedding after 10 days of treatment, antiviral drug resistance testing should be performed. The doses and courses of antiviral drugs should be adjusted on an individual basis (24). In the current study, patient 2 was isolated for more than 7 days and received prolonged treatment with antivirals. We speculated that immunodeficiency might have contributed to prolonged viral shedding in this patient, as has been reported in some studies (24,25). The monitoring of viral shedding may be a useful guide for antiviral treatment. Further studies are needed to determine whether viral shedding can be used to determine the necessary duration of isolation and the course of antiviral treatment.
Health care workers can contribute to nosocomial influenza virus transmission because they can be exposed to influenza in both the community and the hospital (8). These patients may be symptomatic or asymptomatic and might not follow standard isolation precautions or precautions specific to control of the dissemination of respiratory droplets at work. Hence, health care workers are important potential sources of influenza outbreaks in hospital settings. Many reports have described the transmission of influenza from health care workers to patients (14,26,27). Previously published data suggested that health care workers were the index patients in nosocomial influenza outbreaks. However, in our study, there was no sign of acute febrile illness among health care workers. The fact that throat swabs were not obtained from the health care workers and that health care workers were not tested for the presence of the influenza virus should be regarded as major limitations. A number of studies have shown increases in the rates of influenza virus vaccination among health care workers, and such increases were found to be associated with reductions in the incidence of nosocomial influenza infection (28,29).
Antiviral treatment is an effective approach to shorten the duration of viral shedding, reduce the viral load in vivo and alleviate symptoms, provided it can be administered within 48 or 72 hours after illness onset (18,30,31). However, patients with severe influenza or high-risk patients might still benefit from antiviral treatment, even when it is administered 72 hours or more after illness onset (32,33). In our study, oseltamivir was administered to most patients once the diagnosis of influenza was suspected or confirmed. In the present study, prolonged hospital stays and increased costs were major consequences, similar to other studies (9,34). Excess mortality was also reported in the literature (9). However, none of the patients with confirmed cases developed complications, except one patient with bronchiectasis and Penicillium marneffei infection who had a prolonged period of viral shedding, in our study. The patients recovered from influenza after antiviral treatment with oseltamivir. Studies have shown that prolonged shedding of the virus is associated with drug resistance (35,36). However, the common drug resistance mutation H275Y in the NA protein was not identified in the isolates in our study. Antiviral treatment should be prolonged for 7 days or more in patients infected with influenza with this mutation (37). Routine monitoring of the viral load in immunocompromised populations might help determine the course of antiviral treatment.
Both shorter and longer courses of neuraminidase inhibitor chemoprophylaxis were effective in patients with exposure to influenza patients, preventing the nosocomial spread of influenza (38-40). Hence, it is recommended by the Infectious Diseases Society of America to reduce the secondary spread of influenza (18). In the present study, although there were new cases after identification of the index case, the route of transmission from the index patient to the new patients could not be confirmed. Transmission of the influenza virus between these influenza patients was retrospectively confirmed by phylogenetic analysis. Hence, chemoprophylaxis was difficult to carry out for all the patients in the ward in this situation. The infection control bundle played a role in the prevention of secondary influenza and the containment of the nosocomial influenza outbreak in the present study, but the bundle did not include chemoprophylaxis.
There are other limitations that should be considered. First, there was no sampling of the air or surfaces in our ward or of the instruments within the bronchoscopy room; in addition, visitors were not tested for influenza. Therefore, we cannot confirm the source of this outbreak. Second, only four influenza strains were successfully recovered and sequenced. For those from whom isolates were not obtained, three patients tested positive for H1N1 on real-time RT-PCR, and one patient tested negative on the rapid influenza antigen test. It remains unclear whether all eight patients were infected with the same influenza strain.
Conclusions
Clinicians should be vigilant with regard to the risk of nosocomial A(H1N1)pdm09 infection in hospitalized patients. After the implementation of infection control measures, the nosocomial A(H1N1)pdm09 outbreak was rapidly contained; those with fever experienced a delay in discharge and excess costs, but no mortality occurred.
Acknowledgments
All the authors thank all the patients for their contributions and informed consent.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-897
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-897
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-897). Prof. RC serves as the Associate Editor-in-Chief of Journal of Thoracic Disease. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study was approved by the Ethics Committee of Scientific Research Projects of the First Affiliated Hospital of Guangzhou Medical University (No. 2014.14) and informed consent was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou L, Yang H, Kuang Y, et al. Temporal patterns of influenza A subtypes and B lineages across age in a subtropical city, during pre-pandemic, pandemic, and post-pandemic seasons. BMC Infect Dis 2019;19:89. [Crossref] [PubMed]
- Tete SM, Jul-Larsen Å, Rostami S, et al. Impact of pre-existing immunity on the induction of functional cross-reactive anti-hemagglutinin stalk antibodies following vaccination with an AS03 adjuvanted pandemic H1N1 vaccine. Vaccine 2018;36:2213-9. [Crossref] [PubMed]
- Mourmouris P, Tzelves L, Roidi C, et al. COVID-19 transmission: a rapid systematic review of current knowledge. Osong Public Health Res Perspect 2021;12:54-63. [Crossref] [PubMed]
- Ameme DK, Dadzie D, Asiedu-Bekoe F, et al. Influenza A (H1N1)pdm09 outbreak of unknown source in a Ghanaian senior high school. BMC Public Health 2020;20:1423. [Crossref] [PubMed]
- Ninomiya-Mori A, Nukuzuma S, Suga T, et al. Genetic evidence for containment of viruses in the first outbreak of influenza A pandemic (H1N1) 2009 in Kobe, Japan. Influenza Other Respir Viruses 2011;5:180-7. [Crossref] [PubMed]
- Russell ES, Zheteyeva Y, Gao H, et al. Reactive School Closure During Increased Influenza-Like Illness (ILI) Activity in Western Kentucky, 2013: A Field Evaluation of Effect on ILI Incidence and Economic and Social Consequences for Families. Open Forum Infect Dis 2016;3:ofw113 [Crossref] [PubMed]
- Xiao S, Tang JW, Hui DS, et al. Probable transmission routes of the influenza virus in a nosocomial outbreak. Epidemiol Infect 2018;146:1114-22. [Crossref] [PubMed]
- Salgado CD, Farr BM, Hall KK, et al. Influenza in the acute hospital setting. Lancet Infect Dis 2002;2:145-55. [Crossref] [PubMed]
- Yang K, Zhang N, Gao C, et al. Risk factors for hospital-acquired influenza A and patient characteristics: a matched case-control study. BMC Infect Dis 2020;20:863. [Crossref] [PubMed]
- Chen H, Liu S, Liu J, et al. Nosocomial Co-Transmission of Avian Influenza A(H7N9) and A(H1N1)pdm09 Viruses between 2 Patients with Hematologic Disorders. Emerg Infect Dis 2016;22:598-607. [Crossref] [PubMed]
- Fang CF, Ma MJ, Zhan BD, et al. Nosocomial transmission of avian influenza A (H7N9) virus in China: epidemiological investigation. BMJ 2015;351:h5765. [Crossref] [PubMed]
- Zhan Y, Yang Z, Chen R, et al. Respiratory virus is a real pathogen in immunocompetent community-acquired pneumonia: comparing to influenza like illness and volunteer controls. BMC Pulm Med 2014;14:144. [Crossref] [PubMed]
- Ip DK, Lau LL, Leung NH, et al. Viral Shedding and Transmission Potential of Asymptomatic and Paucisymptomatic Influenza Virus Infections in the Community. Clin Infect Dis 2017;64:736-42. [PubMed]
- Eibach D, Casalegno JS, Bouscambert M, et al. Routes of transmission during a nosocomial influenza A(H3N2) outbreak among geriatric patients and healthcare workers. J Hosp Infect 2014;86:188-93. [Crossref] [PubMed]
- Cho SY, Kang JM, Ha YE, et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet 2016;388:994-1001. [Crossref] [PubMed]
- Jung J, Lee J, Jo S, et al. Nosocomial Outbreak of COVID-19 in a Hematologic Ward. Infect Chemother 2021;53:332-41. [Crossref] [PubMed]
- Boender TS, Bender JK, Krüger A, et al. Factors preventing SARS-CoV-2 transmission during unintentional exposure in a GP practice: a cohort study of patient contacts; Germany, 2020. Epidemiol Infect 2021;149:e161 [Crossref] [PubMed]
- Uyeki TM, Bernstein HH, Bradley JS, et al. Clinical Practice Guidelines by the Infectious Diseases Society of America: 2018 Update on Diagnosis, Treatment, Chemoprophylaxis, and Institutional Outbreak Management of Seasonal Influenzaa. Clin Infect Dis 2019;68:e1-e47. [Crossref] [PubMed]
- Centers for Disease Control and Prevention (CDC). Prevention Strategies for Seasonal Influenza in Healthcare Settings. Available online: https://www.cdc.gov/flu/professionals/infectioncontrol/healthcaresettings.htm
- Leekha S, Zitterkopf NL, Espy MJ, et al. Duration of influenza A virus shedding in hospitalized patients and implications for infection control. Infect Control Hosp Epidemiol 2007;28:1071-6. [Crossref] [PubMed]
- Bruminhent J, Deziel PJ, Wotton JT, et al. Prolonged shedding of pandemic influenza A (H1N1) 2009 virus in a pancreas-after-kidney transplant recipient. J Clin Virol 2014;61:302-4. [Crossref] [PubMed]
- Fraaij PL, Schutten M, Javouhey E, et al. Viral shedding and susceptibility to oseltamivir in hospitalized immunocompromised patients with influenza in the Influenza Resistance Information Study (IRIS). Antivir Ther 2015;20:633-42. [Crossref] [PubMed]
- Centers for Disease Control and Prevention (CDC). Interim Guidance for Influenza Outbreak Management in Long-Term Care and Post-Acute Care Facilities. Available online: https://www.cdc.gov/flu/professionals/infectioncontrol/ltc-facility-guidance.htm
- Mitha E, Krivan G, Jacobs F, et al. Safety, Resistance, and Efficacy Results from a Phase IIIb Study of Conventional- and Double-Dose Oseltamivir Regimens for Treatment of Influenza in Immunocompromised Patients. Infect Dis Ther 2019;8:613-26. [Crossref] [PubMed]
- von Mollendorf C, Hellferscee O, Valley-Omar Z, et al. Influenza Viral Shedding in a Prospective Cohort of HIV-Infected and Uninfected Children and Adults in 2 Provinces of South Africa, 2012-2014. J Infect Dis 2018;218:1228-37. [Crossref] [PubMed]
- Wilson KE, Wood SM, Schaecher KE, et al. Nosocomial outbreak of influenza A H3N2 in an inpatient oncology unit related to health care workers presenting to work while ill. Am J Infect Control 2019;47:683-7. [Crossref] [PubMed]
- Cavari Y, Kaplan O, Zander A, et al. Healthcare workers mobile phone usage: A potential risk for viral contamination. Surveillance pilot study. Infect Dis (Lond) 2016;48:432-5. [Crossref] [PubMed]
- Frenzel E, Chemaly RF, Ariza-Heredia E, et al. Association of increased influenza vaccination in health care workers with a reduction in nosocomial influenza infections in cancer patients. Am J Infect Control 2016;44:1016-21. [Crossref] [PubMed]
- Bénet T, Régis C, Voirin N, et al. Influenza vaccination of healthcare workers in acute-care hospitals: a case-control study of its effect on hospital-acquired influenza among patients. BMC Infect Dis 2012;12:30. [Crossref] [PubMed]
- Doll MK, Winters N, Boikos C, et al. Safety and effectiveness of neuraminidase inhibitors for influenza treatment, prophylaxis, and outbreak control: a systematic review of systematic reviews and/or meta-analyses. J Antimicrob Chemother 2017;72:2990-3007. [Crossref] [PubMed]
- Muthuri SG, Venkatesan S, Myles PR, et al. Impact of neuraminidase inhibitors on influenza A(H1N1)pdm09-related pneumonia: an individual participant data meta-analysis. Influenza Other Respir Viruses 2016;10:192-204. [Crossref] [PubMed]
- Louie JK, Yang S, Acosta M, et al. Treatment with neuraminidase inhibitors for critically ill patients with influenza A (H1N1)pdm09. Clin Infect Dis 2012;55:1198-204. [Crossref] [PubMed]
- Yu H, Feng Z, Uyeki TM, et al. Risk factors for severe illness with 2009 pandemic influenza A (H1N1) virus infection in China. Clin Infect Dis 2011;52:457-65. [Crossref] [PubMed]
- Diel R, Nienhaus A. Rapid Point-of-Care Influenza Testing for Patients in German Emergency Rooms - A Cost-Benefit Analysis. J Health Econ Outcomes Res 2019;6:203-12. [Crossref] [PubMed]
- Roosenhoff R, van der Vries E, van der Linden A, et al. Influenza A/H3N2 virus infection in immunocompromised ferrets and emergence of antiviral resistance. PLoS One 2018;13:e0200849 [Crossref] [PubMed]
- Roosenhoff R, Reed V, Kenwright A, et al. Viral Kinetics and Resistance Development in Children Treated with Neuraminidase Inhibitors: The Influenza Resistance Information Study (IRIS). Clin Infect Dis 2020;71:1186-94. [Crossref] [PubMed]
- Centers for Disease Control and Prevention (CDC). Influenza Antiviral Medications: Summary for linnicians,2015. Available online: http://www.cdc.gov/flu/professionals/antivirals/summary-clinicians.htm
- Shinjoh M, Takano Y, Takahashi T, et al. Postexposure prophylaxis for influenza in pediatric wards oseltamivir or zanamivir after rapid antigen detection. Pediatr Infect Dis J 2012;31:1119-23. [Crossref] [PubMed]
- Ishiguro N, Oyamada R, Nasuhara Y, et al. Three-day regimen of oseltamivir for postexposure prophylaxis of influenza in wards. J Hosp Infect 2016;94:150-3. [Crossref] [PubMed]
- Higa F, Tateyama M, Tomishima M, et al. Role of neuraminidase inhibitor chemoprophylaxis in controlling nosocomial influenza: an observational study. Influenza Other Respir Viruses 2012;6:299-303. [Crossref] [PubMed]


