Prognostic value of an immunohistochemical signature in patients with pulmonary squamous cell carcinoma undergoing complete surgical resection
Introduction
Lung cancer is one of the leading causes of death worldwide (1,800,000 deaths per year), with its incidence increasing every year (1). Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for 85–90% of all lung cancer cases. The 2 major histological types of lung cancers are squamous cell carcinoma (SQCC) and adenocarcinoma (2). Radical resection is still the main treatment for SQCC (3). Although Tumor Node Metastasis (TNM) stage can predict the prognosis of patients, the prognosis for high-risk lung cancer patients is not poor (4). And based on TNM stage, the patients with chemotherapy have poor interests in their progression-free survival (5). Therefore, it is necessary to improve postoperative prognosis by integrating multiple biomarker predictive models to guide patient management after surgery.
Microscopic morphological characteristics of tumors have always been the gold standard for lung cancer classification by the World Health Organization. In cases of poor tumor differentiation and in the absence of lung adenocarcinoma and cytological characteristics, classification is more difficult. Due to the need for precise tumor typing, reliance solely on morphology for diagnosis has been challenged in recent years. Because of this, immunohistochemical (IHC) detection is important in NSCLC subtyping. IHC is widely used because it is simple, relatively inexpensive, and highly reliable. In clinical practice, in addition to p63, p40, and CK5/6(Cytokeratins 5/6), there are many epithelial cell markers, including 34BE12, desmocollin-3, S100A2, S100A7, SOX2(SRY-Box Transcription Factor 2), glypican 3, and miR-205, which have been used to identify SQCC. Increasing evidence suggests that IHC is a highly effective aid in predicting survival in patients with various cancer types. For example, high Ki67 has a close association with poor prognosis in colorectal cancer (6). A 3-gene IHC panel has been reported to predict the prognosis of patients with esophageal adenocarcinoma (7). In clinical, we evaluate patients who need chemotherapy based on IHC. Several IHC-based biomarkers have been reported to predict various cancers (8), but there are few reports pertaining to SQCC. The integration of multiple biomarkers into a single predictive model could enable clinicians to optimize patient treatment and reduce mortality.
The purpose of the present study was to develop and validate IHC-based classifiers using the least absolute shrinkage and selection operator (LASSO). Cox regression model and to establish a prognosis for postoperative patients with pulmonary SQCC based on clinical-pathological parameters and IHC biomarkers.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-118/rc).
Methods
Patients and samples
Patients who underwent SQCC resection at Ningbo Medical Center Lihuili Hospital, affiliated with Ningbo University, from January 2010 to December 2014 with complete clinical and immunological data were included in the present study (n=556). Patient selection criteria: a. Initial diagnosis is SQCC; b. The follow-up data is relatively complete; c. Patients diagnosed the disease as a solitary disease.
Patient exclusion criteria: a. Patients with tumor staging deficit; b. Patients with other malignant tumors; c. Patients with lack of progression-free survival; d. Patients with missing follow-up time.
The study was approved by the ethics committee of Ningbo Medical Center Lihuili Hospital (No. KY2019PJ058) and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all patients prior to inclusion. Patients were randomly divided into the discovery group (n=334) and the validation group (n=222) at ratio of 6:4.
Immunohistochemistry
The specimens studied were formalin-fixed and paraffin-enucleated tissue slices. Specimens were first analyzed by the primary pathologist, then independently by 2 practicing pathologists. Specimens were then subjected to IHC using antibodies at the Ningbo Pathology Center. The antibodies used were from Fu Zhou Maixin Biotech and included CK pan, CK7, TTF-1(thyroid transcription factor-1), napsinA, CK5/6, p63, p40, CD56, Syn(synaptophysin), CgA(chromogranin A), and Ki67. All the specimens were fixed with 10% neutral formalin, embedded in paraffin, and stained with hematoxylin eosin. IHC staining was performed with EnVision(Dako, Santa Clara, CA, USA). The percentage of positive tumor cells and the maximum intensity of IHC signal [0–3] were recorded.
Development and validation of IHC markers
The LASSO Cox regression model was used. Significant prognostic markers with non-zero coefficients were determined from the test group. The prognostic score for each patient was calculated using a linear combination of the identified markers. In the test group, a multimarker classifier to predict 3- and 5-year progression-free survival (PFS) in SQCC patients was established. The LASSO Cox regression model analysis was performed using the “glmnet” function in R software 3.0.1 (R Foundation for Statistical Computing, Vienna, Austria).
Statistical analysis
We compared the 2 datasets using the t-test for continuous variables and the χ2-test for categorical variables. The Kaplan-Meier survival analysis and log-rank were used to estimate the survival time of patients in the different risk groups stratified according to IHC markers. A quantile plot was used to select the best cut-off point for patient survival time.
Receiver-operating characteristic curve analysis was performed, statistically significant prognostic markers were analyzed using univariate and multivariate Cox regressions, and Cox regression coefficients were used to build a column chart to predict PFS probability. Based on the regression analysis, a calibration plot was developed. We evaluated the clinical utility of the charts through decision curve analysis (DCA). R’s “rms” function was used for line and calibrations plots. Two-tailed P value was 0.05.
Results
Patients’ clinical characteristics
All patients with SQCC underwent surgery resection and had histologically negative resection margins. The median follow-up time was 36 months (range, 1–116 months), during which there were 263 relapses and 235 deaths. The clinical stage of patients was determined according to the TNM staging of lung cancer in the 8th edition of the NICC (9). Detailed clinicopathological characteristics of the discovery group (n=334) and the validation group (n=222) are shown in Table 1.
Table 1
| Characteristics | Training set | Testing set | |||
|---|---|---|---|---|---|
| low risk (n=132) | high risk (n=202) | low risk (n=90) | high risk (n=132) | ||
| Age, n (%) | |||||
| ≤60 years | 43 (32.6) | 66 (32.7) | 31 (34.4) | 46 (34.8) | |
| >60 years | 89 (67.4) | 136 (67.3) | 59 (65.6) | 86 (65.2) | |
| Smoker, n (%) | |||||
| No | 47 (35.6) | 57 (28.2) | 38 (42.2) | 42 (31.8) | |
| Yes | 85 (64.4) | 145 (71.8) | 52 (57.8) | 90 (68.2) | |
| Sex, n (%) | |||||
| Female | 4 (3.0) | 9 (4.5) | 7 (7.8) | 10 (7.6) | |
| Male | 128 (97.0) | 193 (95.5) | 83 (92.2) | 122 (92.4) | |
| Lymphatic or blood vessel invasion, n (%) | |||||
| No | 110 (83.3) | 157 (77.7) | 76 (84.4) | 107 (81.1) | |
| Yes | 22 (16.7) | 45 (22.3) | 14 (15.6) | 25 (18.9) | |
| Nerve invasion, n (%) | |||||
| No | 115 (87.1) | 174 (86.1) | 82 (91.1) | 121 (91.7) | |
| Yes | 17 (12.9) | 28 (13.9) | 8 (8.9) | 11 (8.3) | |
| Pleural invasion, n (%) | |||||
| No | 118 (89.4) | 159 (78.7) | 86 (95.6) | 115 (87.1) | |
| Yes | 14 (10.6) | 43 (21.3) | 4 (4.4) | 17 (12.9) | |
| Differentiation, n (%) | |||||
| W | 21 (15.9) | 22 (10.9) | 10 (11.1) | 13 (9.8) | |
| L | 47 (35.6) | 82 (40.6) | 34 (37.8) | 50 (37.9) | |
| M | 64 (48.5) | 98 (48.5) | 46 (51.1) | 69 (52.3) | |
| Stage n (%) | |||||
| I | 76 (57.6) | 64 (31.7) | 55 (61.1) | 55 (41.7) | |
| II | 36 (27.3) | 55 (27.2) | 21 (23.3) | 33 (25.0) | |
| III | 20 (15.2) | 83 (41.1) | 14 (15.6) | 44 (33.3) | |
Selection and grouping of IHC markers
The LASSO Cox regression model was used to identify potential predictive IHC markers (SOX2, CEA (carcinoembryonic antigen), Ki67, p53, CK5/6, 34BE12, p63, CD44V6, COX-2(cyclooxygenase-2), PGP-9.5(Protein gene product 9.5), EGFR(epidermal growth factor receptor), VEGF(vascular endothelial growth factor), c-Myc, BCL-2(B-cell lymphoma-2), Topo-II(Type II topoisomerases), cyclin D1, p16, p21, p27) (Figure 1). We used the LASSO method to reduce the regression coefficient and reduce 19 characteristics to 4 potential predictors: SOX2, VEGF, p63, and p27. According to the expression status of the four IHC markers, we derived a formula to calculate the risk score of individual patients, based on their individual four prognostic marker expression levels: Risk score = 0.063372939 × SOX2 – 0.004970944 × p63 + 0.106611323 × VEGF – 0.013048563 × p27. In this formula, the negative status of IHC equals 0 and positive status equals 1. The optimum cut-off level of 6 markers was defined as 0.10 by the X-tile plot approach (Figure S1). Consequently, using this formula, patients with scores greater than or equal to 0 (n=202, 60.5%) were classified as high risk; those with scores less than 0 (n=132, 39.5%) were classified as low risk (Figure 2). PFS was significantly better in the low-risk group.


Validation of significance
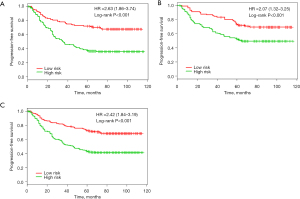
There were significant differences in survival rates among patients in the different risk groups. PFS was significantly better in the low-risk subgroups than in the high-risk subgroups in the discovery group, the validation group, and the combined cohort [P<0.001, HR =2.63 (95% CI, 1.86–3.74)]. The same analysis was carried out in the validation group (n=222) [P<0.001, HR =2.07 (95% CI, 1.32–3.25)]. Using the risk score, we classified these patients into a high-risk group (n=132, 59.5%) and a low-risk group (n=90, 40.5%). Similar differences between the 2 groups were noted in the combined discovery and validation cohort [P<0.001, HR =2.42 (95% CI, 1.84–3.19)] (Figure 3).
Prediction accuracy
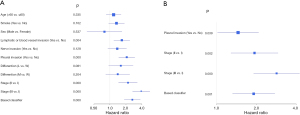
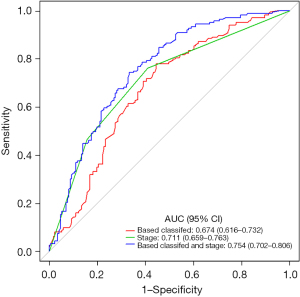
Pleural invasion, staging, and IHC-based classifiers were significant prognostic factors in the univariate analysis (Figure 4A). There was no statistically significant difference in other clinicopathological factors. The multivariate analysis identified tumor-stage markers and IHC markers to be independent PFS predictors (Figure 4B). Furthermore, combining IHC-based classifiers with staging provided better predictive value than IHC-based classifiers or staging alone (P<0.01) (Figure 5). Therefore, IHC-based classifiers can increase the prognostic value of staging in post-resection SQCC patients.
Nomogram and clinical application
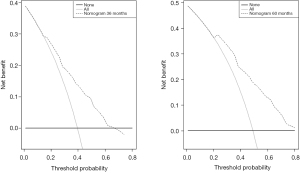
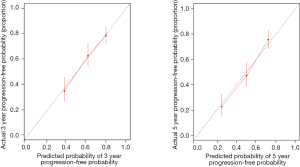
To provide a clinically relevant tool for predicting prognosis, we built a nomogram that integrates a variety of clinicopathological risk factors based on IHC-based classifiers and PFS (Figure 6). Predictive factors included pleural invasion, tumor stage, and IHC-based classifiers. The calibration curve showed optimal performance of the nomogram with a high degree of consistency between the PFS and the Kaplan-Meier estimates (Figure 7).
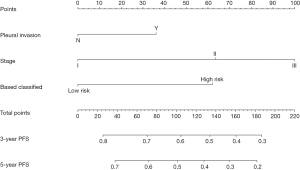
DCA was used to evaluate the clinical utility of a nomogram based on IHC-based classifiers by quantifying net benefits (Figure 8). The threshold probability of a patient choosing treatment tells us how the patient weighs the relative harm of false-positive and false-negative predictions. Here, the relative harm from treatment is equal to that of avoiding the expected benefits of treatment. Net benefit was calculated by subtracting the proportion of false positives from the sum of false and true positives. The nomogram proved to be of clinical value, as it ensured a better net benefit in comparison to the full or no treatment option.
Discussion
Surgical resection is the most effective treatment for lung cancer, but predicting postoperative patients’ likelihood of recurrence or PFS is difficult (10). The current method does not provide viable clinical prognostic markers in postoperative patients with SQCC (11). To help guide decision making in the management of SQCC patients, we used a group of SQCC patients to analyze IHC markers that were associated with prognosis and developed a nomogram to estimate 3- and 5-year PFS in postoperative patients.
A LASSO Cox regression was used to identify statistically significant prognostic factors; this regression builds models based on both predictors and selected characteristics. In a recent study, the LASSO Cox regression method was used in multivariate analysis panels, such as preoperative prediction of lymph node metastasis in colorectal cancer (12). Our study used the LASSO method to reduce the regression coefficient and reduce 19 characteristics to 4 potential predictors. Patients were divided into low-risk and high-risk groups by incorporating 4 IHC markers into the classifier. The survival rate of patients in the low-risk group was significantly higher than that in the high-risk group. In addition, we validated the potential value of IHC markers in predicting patient prognosis using the validation group. Multivariate analysis showed that IHC markers are independent prognostic factors of PFS after adjustment is made for clinicopathological variables. Combined with staging, IHC markers provide better predictive value than staging alone. IHC and clinicopathological variables, including invasion status and staging, were included in the nomogram. The calibration graph showed significant correlation between the predicted survival probability and the observed survival rate. DCA showed great potential for clinical applicability of the nomogram.
IHC technology has advanced from traditional cell microscopic examination technology. It provides a better understanding of enzyme activity and the shape and structure of the patient’s cells, as well as a more accurate understanding of the histopathology of different lesions (13). It is highly accurate and specific. SOX2 is an important member of the SOX family, which regulates the development of embryos and tissues by binding to the HMG (high mobility group) domain of the target gene. It maintains the pluripotent, undifferentiated state of the stem cells, regulates their proliferation, and determines their cell fate (14). Numerous studies have found SOX2 expression abnormalities in many solid tumors, such as breast cancer and malignant gliomas (15). One study found that amplification of SOX2 loci can be detected in early stage of NSCLC, which is associated with median overall survival in such cohort, but not found in patients with advanced NSCLC (16). Another study showed that the prognosis of tumor patients with a high expression of SOX2 was significantly better than that of patients with a low expression (17). Interestingly, Chou and his colleagues reported that the expression of SOX2 was associated with the staging of NSCLC (18). For lymph node metastasis and TNM stage, the lower the degree of tumor differentiation, the earlier the metastasis to the lymph nodes; the later the TNM stage, the higher the expression rate of SOX2 (19). This suggests that the expression of SOX2 might be closely related to the proliferation and metastasis process of NSCLC and a relevant factor in poor tumor prognosis. Therefore, it is proposed that SOX2 expression can be used to predict NSCLC.
The human p63 gene, located on chromosomes 3q27–3q29, consists of 15 exons that each consist of 2 independent initiators. At present, the p63 (monoclonal antibody cloning system 4A4) used in domestic and foreign pathology laboratories can identify TAp63 and Xenon Np63, and it is considered to be a “broad-spectrum” p63 antibody, while the p40 antibody Np63 only recognizes p-Np63 (20). The p63 protein is often expressed at the base of the epithelial tissue, playing an important role in the formation of normal endothelial tissue. It is expressed in a variety of malignant tumors of squamous cell origin, especially in SQCC tissue. Its frequency and expression distribution are related to the degree of SQCC differentiation (21). In breast cancer, the expression of p63 has been demonstrated to be related to tumor grading and is significant for the prognosis and treatment index of tumors (22).
The p27Kip1 protein is a cell cycle inhibitor, discovered in a 1994, which was used to study of cell suppression mechanisms. Studies have found that p27Kip1 is reduced or missing in a variety of malignant tumor tissues, including esophageal, colon, prostate, bladder, liver, breast, and NSCLC (23-29). The expression of p27, as identified via reverse transcription polymerase chain reaction (RT-PCR), was significantly lower than that in adjacent paracancerous tissues in 230 cases of NSCLC, there were significant differences (P<0.050) in lymph node metastasis, TNM staging, and differentiation classification (30).
VEGF is the most consequential cytokine known to promote the growth of the endothelium of blood vessels. It can increase vascular permeability, promote tumor vascular formation, and plays an important role in the proliferation and migration of tumor vascular endothelial cells. In vitro animal model experiments have shown that the inhibition of VEGF expression in tumors can significantly inhibit the formation of blood vessels in lung cancer cell tissue, reduce tumor volume, and improve survival time in mice (29). Other studies have shown that VEGF increases significantly in NSCLC patients, and its elevation has been found to be associated with tumor TNM stage and lymph node metastasis (31). It was suggested in a meta-analysis that VEGF increases the risk of lung cancer deterioration, could be involved in the leaching metastasis of NSCLC, and accelerates the malignant progression of tumors (32).
In the present study, we used the LASSO Cox regression model to successfully integrate multiple IHC markers into a single model that had higher predictive power than a single IHC marker. Although IHC marker-based nomograms can accurately predict the survival of patients with lung squamous cancer, there are some limitations to our study. First, the nomogram was based on retrospective data from a single center. Second, our study lacked data on the genetic characteristics needed to validate the biomarkers. Third, further prospective studies of multicenter clinical trials are needed to further validate the results. In summary, we developed and validated a nomogram combining IHC markers and clinicopathological characteristics to accurately predict the prognosis of patients with SQCC. An accurate prognostic model for predicting patient survival can be of significant value in choosing the best treatment strategy and individualizing patient care.
Acknowledgments
The authors thank Ningbo Pathology Center for the assistance the study.
Funding: This work was supported by the Major Science and Technology Innovation in 2025 Projects of Ningbo, China (2019B10039) and Ningbo Clinical Research Center for Thoracic & Breast Neoplasms (2021L002).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-118/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-118/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-118/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethics committee of Ningbo Medical Center Lihuili Hospital (No. KY2019PJ058) and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all patients prior to inclusion.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Youlden DR, Cramb SM, Baade PD. The International Epidemiology of Lung Cancer: geographical distribution and secular trends. J Thorac Oncol 2008;3:819-31. [Crossref] [PubMed]
- Datta D, Lahiri B. Preoperative evaluation of patients undergoing lung resection surgery. Chest 2003;123:2096-103. [Crossref] [PubMed]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. [Crossref] [PubMed]
- Rossi A, Di Maio M. Platinum-based chemotherapy in advanced non-small-cell lung cancer: optimal number of treatment cycles. Expert Rev Anticancer Ther. 2016;16:653-60. [Crossref] [PubMed]
- Tong G, Zhang G, Liu J, et al. Cutoff of 25% for Ki67 expression is a good classification tool for prognosis in colorectal cancer in the AJCC-8 stratification. Oncol Rep 2020;43:1187-98. [Crossref] [PubMed]
- Ong CA, Shapiro J, Nason KS, et al. Three-gene immunohistochemical panel adds to clinical staging algorithms to predict prognosis for patients with esophageal adenocarcinoma. J Clin Oncol 2013;31:1576-82. [Crossref] [PubMed]
- Cagle PT, Allen TC, Olsen RJ. Lung cancer biomarkers: present status and future developments. Arch Pathol Lab Med 2013;137:1191-8. [Crossref] [PubMed]
- Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: External Validation of the Revision of the TNM Stage Groupings in the Eighth Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 2017;12:1109-21.
- Taylor MD, Nagji AS, Bhamidipati CM, et al. Tumor recurrence after complete resection for non-small cell lung cancer. Ann Thorac Surg 2012;93:1813-20; discussion 1820-1. [Crossref] [PubMed]
- Xi JJ, Yin JC, Wang L, et al. A surveillance method-oriented detection of post-operative spatial-temporal recurrence for non-small cell lung cancer. J Thorac Dis 2018;10:6107-17. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Jain D, Nambirajan A, Borczuk A, et al. Immunocytochemistry for predictive biomarker testing in lung cancer cytology. Cancer Cytopathol 2019;127:325-39. [Crossref] [PubMed]
- Novak D, Hüser L, Elton JJ, et al. SOX2 in development and cancer biology. Semin Cancer Biol 2020;67:74-82. [Crossref] [PubMed]
- Gangemi RM, Griffero F, Marubbi D, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 2009;27:40-8. [Crossref] [PubMed]
- Toschi L, Finocchiaro G, Nguyen TT, et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PLoS One 2014;9:e95303. [Crossref] [PubMed]
- Ying J, Shi C, Li CS, et al. Expression and significance of SOX2 in non-small cell lung carcinoma. Oncol Lett 2016;12:3195-8. [Crossref] [PubMed]
- Chou YT, Lee CC, Hsiao SH, et al. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling in lung cancer. Stem Cells 2013;31:2607-19. [Crossref] [PubMed]
- Iijima Y, Seike M, Noro R, et al. Prognostic significance of PIK3CA and SOX2 in Asian patients with lung squamous cell carcinoma. Int J Oncol 2015;46:505-12. [Crossref] [PubMed]
- Bishop JA, Teruya-Feldstein J, Westra WH, et al. p40 (ΔNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol 2012;25:405-15. [Crossref] [PubMed]
- Saghravanian N, Anvari K, Ghazi N, et al. Expression of p63 and CD44 in oral squamous cell carcinoma and correlation with clinicopathological parameters. Arch Oral Biol 2017;82:160-5. [Crossref] [PubMed]
- Constantinou C, Papadopoulos S, Karyda E, et al. Expression and Clinical Significance of Claudin-7, PDL-1, PTEN, c-Kit, c-Met, c-Myc, ALK, CK5/6, CK17, p53, EGFR, Ki67, p63 in Triple-negative Breast Cancer-A Single Centre Prospective Observational Study. In Vivo 2018;32:303-11. [PubMed]
- Shibata H, Matsubara O, Wakiyama H, et al. The role of cyclin-dependent kinase inhibitor p27 in squamous cell carcinoma of the esophagus. Pathol Res Pract 2001;197:157-64. [Crossref] [PubMed]
- Dai JY, Liang XP, Wen JL, et al. Expression of P27 protein and cyclin E in colon cancer. Ai Zheng 2003;22:1093-5. [PubMed]
- Ribal MJ, Fernandez PL, Lopez-Guillermo A, et al. Low p27 expression predicts biochemical relapse after radical prostatectomy in patients with clinically localised prostate cancer. Anticancer Res 2003;23:5101-6. [PubMed]
- Lacoste-Collin L, Gomez-Brouchet A, Escourrou G, et al. Expression of p27(Kip1) in bladder cancers: immunohistochemical study and prognostic value in a series of 95 cases. Cancer Lett 2002;186:115-20. [Crossref] [PubMed]
- Lei PP, Zhang ZJ, Shen LJ, et al. Expression and hypermethylation of p27 kip1 in hepatocarcinogenesis. World J Gastroenterol 2005;11:4587-91. [Crossref] [PubMed]
- Sgambato A, Zhang YJ, Arber N, et al. Deregulated expression of p27(Kip1) in human breast cancers. Clin Cancer Res 1997;3:1879-87. [PubMed]
- Huang H, Salavaggione O, Rivera L, et al. Woodchuck VEGF (wVEGF) characteristics: Model for angiogenesis and human hepatocellular carcinoma directed therapies. Arch Biochem Biophys 2019;661:97-106. [Crossref] [PubMed]
- Yang L, Yang J. Expression and clinical significance of microRNA-21, PTEN and p27 in cancer tissues of patients with non-small cell lung cancer. Oncol Lett 2020;20:49. [Crossref] [PubMed]
- Zheng CL, Qiu C, Shen MX, et al. Prognostic impact of elevation of vascular endothelial growth factor family expression in patients with non-small cell lung cancer: an updated meta-analysis. Asian Pac J Cancer Prev 2015;16:1881-95. [Crossref] [PubMed]
- Fan J, Zhang W, Lei C, et al. Vascular endothelial growth factor polymorphisms and lung cancer risk. Int J Clin Exp Med 2015;8:6406-11. [PubMed]
(English Language Editor: R. Scott)