
Prognostic significance of pyroptosis-related factors in lung adenocarcinoma
Introduction
Lung cancer (LC) is one of the most common neoplasia and the leading cause of worldwide cancer-related lethality (1). Lung adenocarcinoma (LUAD) comprises the largest proportion in LC, accounting for 40% of all cases, with a survival rate of only 4–17% (2). Canonical first-line treatments for LUAD including surgery, chemotherapy, and radiotherapy along with recent popular target agents and immunotherapy (3). Nevertheless, despite the use of multiple therapeutic modalities for the past several decades, the improvement in survival duration for LUAD has remained unfavorable (4). It is necessary to identify a new series of biomarkers that could help to foresee the LUAD prognosis, especially in the level of multi-gene signatures. Pyroptosis has become one of the frontier hotspots of tumor therapy over recent years (5). Research has demonstrated that high expression of gasdermin D (GSDMD), a crucial pyroptotic effector, independently indicates a poor outcome in LUAD (6). Based on the former discovery, we postulated that a set of pyroptotic gene signatures might deliver deeper insight into the prognostic value of LUAD.
Pyroptosis is an inflammatory mode of regulated cell death (RCD) (7-9). It presents pore-forming morphological characteristics such as cell swelling, plasma membrane lysis, chromatin condensation, DNA fragmentation, and intact nucleus, with subsequent release of damage-associated molecular patterns (DAMPs) and pro-inflammatory factors such as interleukin (IL)-1β/18 (10). Although apoptosis and pyroptosis are both processes of RCD, apoptosis is a non-inflammatory mode with intact and packed morphological features (11). The gasdermin family is the essential member of the pyroptotic pathway, where activated gasdermin-combined lipid membrane oligomers slot pore channels and trigger pyroptosis. It has been shown that GSDMD is cleaved by caspase-1/4/5/11 and exposes pore-forming gasdermin N-terminal domain (GSDM-NT) in the inflammasome pathway (12-14). Certain stimulative signals induce non-inflammatory pyroptosis by cleavage of GSDME via activated caspase-3 (15,16). Pyroptosis plays a bidirectional reaction in tumorigenesis interestingly. While some research discovered the anti-tumor effect of pyroptotic mechanism in other tumors, a few researches announced the promotive action of pyroptosis in the development of LUAD. For example, GSDMD, one of crucial members of pyroptosis, was clarified to promote the development of LUAD via phosphoinositide 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway (6). However, a single biomarker of gasdermins cannot reveal the effective prediction, exposing that to discover a pyroptosis-related gene signature label is needed instead. Some previous studies have reported pyroptosis-related hub genes and raised their prognostic models (17-19). Here, we observed a wider range of pyroptosis-related genes that could be included in the screening scope and utilized the methods with correlation to prognostic detectable.
Pyroptosis acts as a bridge between tumor-infiltrating immune cell (TIIC) and tumor tissues. Specifically, the discharge of pyroptosis-derived cytokines, especially IL-1β and IL-18, not only alters the TIIC microenvironment and promote the tumorigenesis by escaping immune surveillance, but also can also triggers TIIC recruitment and help immune therapies (20). From this point, we also made an exploratory of TIIC estimation based on pyroptosis-related gene signature in this study.
In this study, we investigated the key biomarkers and pathways that may provide a deeper perspective on the molecular mechanism of LUAD. We also observed pyroptosis-related genes contributing to prognosis in LUAD, and a clinical model of multi-candidate biomarkers was established for anticipating dependable prognosis for LUAD. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-86/rc).
Methods
Data source
We obtained gene expression data and corresponding clinical information of 519 cases of LUAD and 58 cases of normal samples from The Cancer Genome Atlas (TCGA; https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) database to use as a training set. Among them, 490 LUAD samples with complete survival information were involved in the subsequent survival analysis. The GSE31210 dataset (226 LUAD samples in total; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31210) obtained from the Gene Expression Omnibus (GEO) database was used as an external verification set to test the validity of gene tags. The GeneCards database (https://www.genecards.org/) was used to find 110 pyroptosis-related genes (Table S1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential expression analysis
First, we extracted 110 pyroptosis-related gene expression matrices from TCGA expression profile data. Next, the DESeq2 package in R (https://www.r-project.org/) was used for screening the pyroptosis-related differentially expressed genes (DEGs) between the LUAD and normal groups. We set |log2 fold change (FC)| >1 and false discovery rate (FDR) <0.05 as the cut-off criteria to indicate significant statistical difference. In this study, 98 of 110 pyroptosis-related genes were expressed in TCGA-LUAD. The ggplot2 and pheatmap packages in R were used to plot volcano maps and cluster heatmaps.
Prognostic signature construction and validation
To investigate the potential role of pyroptosis-related DEGs in LUAD survival, each pyroptosis-related DEG was assessed by univariate Cox regression analysis and stepwise multivariate Cox proportional hazards regression model in the TCGA-LUAD cohort. Finally, genes with P<0.05 were utilized to construct a prognostic signature and calculate the risk score of each sample. The polygenic risk score was calculated by using following formula: risk score = expression of Gene1 × β1Gene1 + expression of Gene2 × β2Gene2 + … + expression of Genen × βnGenen. Here, the step multivariate Cox proportional hazards regression model was used to generate β, which represented the regression coefficient. The accuracy of the prognostic signature was evaluated by employing the time-dependent receiver operating characteristic (tdROC) curve, which was implemented in the SurvivorROC package. To verify this prognostic signature, LUAD patients from the GSE31210 dataset were treated as an independent external verification cohort.
Independent prognostic analysis
To assess values of independent prediction for four-gene signature in LUAD, univariate and multivariate Cox regression analyses were performed. In this study, age, gender, tumor stage, and pathological tumor, node, metastasis (TNM) stage were included in this analysis, which were obtained from the TCGA-LUAD cohort.
Gene set enrichment analysis (GSEA)
To explore potential mechanisms underlying correlation between prognostic signature and LUAD, GSEA was conducted between high- and low-risk groups in the TCGA cohort through clusterProfiler package in R. The c5 reference gene set and c2 reference gene set from Molecular Signatures Database (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb/) were used as the Gene Ontology (GO) gene set and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set, respectively. By running GSEA, normalized enrichment scores and P values were created. A P value <0.05 was considered statistically significant. In GO analysis, we only focused on the consequences of the biological process (BP) category. The detailed results of the cellular component (CC) and molecular function (MF) were also derived (tables available at https://cdn.amegroups.cn/static/public/jtd-22-86-1.xlsx; https://cdn.amegroups.cn/static/public/jtd-22-86-2.xlsx).
Evaluation of immune cell type components
To evaluate the various abundance of 22 TIIC types between low- and high-risk of LUAD cases, the Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) analytical tool with the LM22 signature matrix was performed. We then measured the P value for the deconvolution of each specimen through Monte Carlo sampling. Cases were enrolled with CIBERSORT P<0.05, the value of which controlled taking in and out (table available at https://cdn.amegroups.cn/static/public/jtd-22-86-3.xlsx). All values of the estimated 22 TIIC proportions always summed up to 1 for every sample.
Statistical analysis
The independent sample t-test was utilized to compare the test of expression between both groups. The DEG screening was adjusted by FDR through the Benjamini-Hochberg methods. Survival analysis performed with the Kaplan-Meier (K-M) method was checked by the log-rank test. A P value <0.05 was considered to achieve statistical significance. Statistical analyses were conducted using the R software.
Results
Identification of DE-pyroptosis-related genes in LUAD
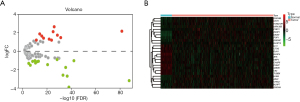
Following TCGA-LUAD database analysis in the DESeq2 package, 26 LUAD-associated DE-pyroptosis-related genes were successfully identified. Notably, 15 pyroptosis-related genes were markedly downregulated, while 11 genes were upregulated in LUAD tissues contrasted to normal counterparts (Figure 1A; Table S2). The hierarchical clustering heatmap of the above 26 genes is shown in Figure 1B, where the red and green dots represent the up and downregulated genes, respectively. The 26 genes obtained from the differential analysis were included as candidate genes for further analysis.
Screening of feature genes for the prognosis of LUAD
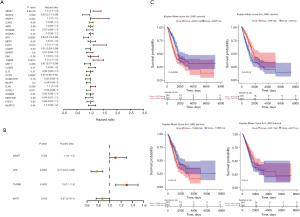
Univariate Cox regression analysis was performed to determine the prognostic value of candidate genes. It was revealed that 10 of 26 candidate genes were significantly associated with OS in LUAD. The MKI67, PARP1, POP1, TUBB6, and GJA1 genes were considered as risk factors [hazard ratio (HR) >1], while the remaining 5 prognostic candidate genes (NLRC4, BTK, CAMP, MST1, and CTSG) were identified as protective factors (HR <1; Figure 2A). Stepwise multivariate Cox regression analysis was applied to define the stable markers, which would build a clinical survival prognostic model, from 10 OS-related candidate genes. The 4 parameters for building the prognostic model are presented in Figure 2B, and 2 filter markers (MKI67 and TUBB6) were shown to be associated with increased risk (HR >1; Table S3). Subsequently, K-M curve analysis was employed for each gene, with the median value of expression as the cut-off (Figure 2C). Here, the expression of MST1 was not significantly associated with OS (P=0.24), and the significance of which was not obvious in formal analysis (P=0.055). After comprehensive consideration, we continued to use it as a key prognostic gene to construct a prognostic model with MKI67, BTK1, and TUBB6. Ultimately, a risk score formula indicating survival prediction for each sample was developed based on the 4 key genes, along with their coefficients and expression levels, which was presented as follows: (0.13 × expression level of MKI67) + (−0.27 × expression level of BTK) + (0.23 × expression level of TUBB6) + (−0.14 × expression level of MST1).
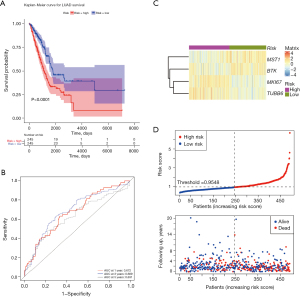
Evaluation and verification of the value of the prognostic signature
To validate the robustness of the 4 pyroptosis-related genes, the capability of the prognostic model to stratify the high- or low-risk group in the TCGA-LUAD dataset was evaluated. A total of 490 patients with LUAD were bisected into a high- or low-risk group with the standard of the median value of the risk score. It showed that the high-risk group by this scoring system was greatly distinguishable from the low-risk group and was linked with the poorer prognosis from the K-M curve analysis (Figure 3A). The 1-, 3-, and 5-year area under the curve (AUC) of the tdROC was 0.672, 0.689, and 0.631, respectively (Figure 3B), corroborating the tolerable prediction efficiency of the 4-gene signature in OS. Consecutively, as exhibited in the heatmap, the expression of MST1 and BTK increased in the low-risk group, while MKI67 and TUBB6 were overexpressed in the high-risk group (Figure 3C). In succession, it was indicated that patients with a risk score of 0.9548 or higher generally survived worse than the other group of patients viewing from the distribution of risk scores and survival status (Figure 3D).
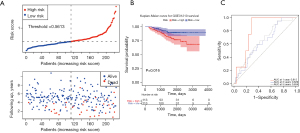
Furthermore, verification of the prognostic capacity of the promising marker in the GSE31210 dataset followed. Survival information and corresponding gene expression data of 226 LUAD patients from the validation set were gathered and imported into the same formula to calculate the risk score of each patient. Using the median risk score as a cut-off value (0.5613), the LUAD samples in the validation set were divided into high- (n=113) and low-risk (n=113) groups, and the survival status and 4-gene expression manners of the 2 groups were compared (Figure 4A,4B). A similar conclusion made by the training set was confirmed: the high-risk group had poorer OS. As shown in Figure 4C, a good sensitivity of the risk score algorithm was certified with the AUCs of the 4-gene signature corresponding to 1, 3, and 5 years of OS were 0.815, 0.629, and 0.654, respectively.
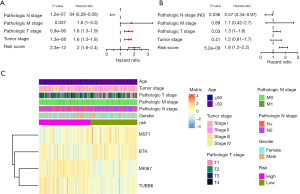
Risk score and pathologic T and N stage as independent prognostic factors for LUAD
Given the independent prognostic value of the 4 pyroptosis-related gene signature, the risk score itself was identified through the univariate and multivariate Cox regression analysis. The TCGA-LUAD dataset suggested that the risk score, pathological TNM stage, and tumor stage were prognostic elements associated with OS (all P<0.05), while age and gender were not associated with the outcome indicator, as evidenced by the forest map of the univariate Cox regression (Figure 5A; Table S4). In sequence, the risk score, pathologic T stage, and pathologic N stage were the chosen potential factors in multivariable Cox regression analyses (Figure 5B; Table S5). The heatmap showed the expression levels of the 4-gene signature and the distribution of clinicopathological features between low- and high-risk TCGA-LUAD patients (Figure 5C). These statistical operations ascertained that the risk score based on the 4-gene signature could be an independent predictive factor for LUAD patients.
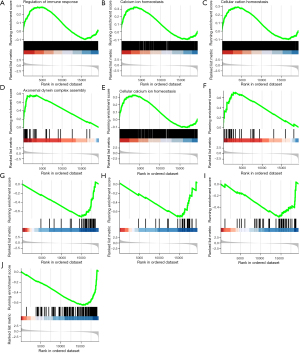
Enriching multiple tumor-associated pathways of the risk group system
In consideration of the risk score derived from the 4-gene signature in connection with some important signaling pathways, GSEA analysis was applied to uncover the relating pathways. The GSEA-GO enrichment results demonstrated that immune response, cell cycle, vasculature development, response to oxidative stress, DNA repair, and so on, were significantly enriched (Figure 6A-6E; table available at https://cdn.amegroups.cn/static/public/jtd-22-86-4.xlsx). Consistent with the GO analysis, the GSEA-KEGG results suggested that genes were involved in the DNA replication, mismatch repair, and cell cycle pathways (Figure 6F-6J). Besides, the risk group system was accompanied by the p53 signaling pathway, cAMP signaling pathway, ether lipid metabolism, peroxisome proliferator-activated receptor (PPAR) signaling pathway, fatty acid degradation, porphyrin and chlorophyll metabolism, and so on (table available at https://cdn.amegroups.cn/static/public/jtd-22-86-5.xlsx). These pathways were significantly correlated with tumor progression.
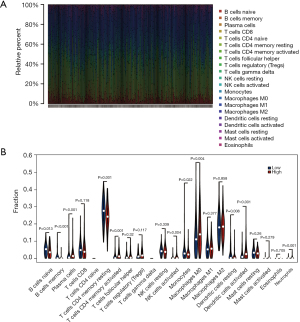
Immune landscapes between LUAD patients with low- and high-risk scores
The exploratory finding was conducted through CIBERSORT, where the variance concerning the scales of 22 TIICs between low- and high-risk LUAD patients was assessed. The proportion of immune cells in each LUAD case was diverse, which indicated that the altered immune cells within the tumor may be an inherent feature representing individual variation (Figure 7A). The high-risk LUAD group contained expressively higher ratios of B cells memory, plasma cells, T cells CD4 memory activated, natural killer (NK) cells activated, macrophages M0, dendritic cells (DCs) activated, and neutrophils (P<0.05), while significantly lower proportions of B cells naive, T cells CD4 memory resting, monocytes, and DC resting than the low-risk LUAD group (Figure 7B). Consequently, the risk score system may cooperate with the heterogeneity of the immune tumor microenvironment (TME) in LUAD to have a critical impact on the identification of prognosis and had significant clinical implications.
Discussion
Present state of diagnosis and therapy in LUAD
Combined strategies of surgery, chemotherapy, radiotherapy, targeted agents, and recently immunotherapy are extensively performed in LUAD (3). Unsatisfactorily, the effectiveness of therapy is less remarkable in advanced LUAD due to the acquired resistance (21,22). In another aspect, LUAD performs more distinctive genomic alterations than other LC types and is accessible to present molecular heterogeneity by genome sequencing technology (23,24). Therefore, genetic mutation-based analysis is supposed to facilitate the therapeutic decisions in improving prognosis.
Connection of pyroptosis, pyroptosis-related hub genes, and immune microenvironment to LUAD
Pyroptosis is a pro-inflammatory manifestation of programmed cell death, traditionally seen as a protective mechanism by self-adjustment or treatments. In this study, we mainly focused on the enriched BP of the 4-gene signature tributed to LUAD, such as immune response, cell cycle, vasculature development, oxidative stress, and DNA repair.
Canonical pyroptosis induces downstream innate and adaptive immune reactions which progressively enhance anti-tumor effect via releasing intracellular molecules (25). Compound L61H10 simultaneously induces G2/M cell cycle arrest and transformation from apoptosis to pyroptosis (26). The protein GSDMD obstructs gastric cancer proliferation by inhibition of CyclinA2 and CDK2 with contributing to S to G2/M phase arrest (27). Piperlongumine analogue L50377 promotes pyroptosis via inducing reactive oxygen species (ROS) generation, which regulates the angiogenesis process (28,29). The ZDHHC1 gene contributes to the increment of oxidative stress to promote pyroptosis for anticancer purposes (30). Additionally, PLK1 inhibitor sensitizes cisplatin-treated DNA damage by negative regulation to DNA repair pathway and induces GSDME-mediated pyroptosis (31).
The pyroptosis-related 4-gene candidates (MKI67, BTK, TUBB6, and MST1) were combined to indicate the prognosis of LUAD in the present filter of the pre-clinical method. The MKI67 gene (marker of proliferation Ki-67) encodes a nuclear DNA-binding protein, Ki-67, which acts as a mitotic chromosome stabilizer by linking to and covering its surface (32). Thus, it has been widely used as a technological marker for cell proliferation in many diseases (33). Ki-67 becomes the same role in the tumorigenesis of LUAD and its overexpression indicates declining OS in LUAD (34). It has been revealed as a poor indication in LUAD, and one of the influential factors in differentiation between LUAD subtypes (35). Simvastatin has been reported to inhibit the effect of Ki-67 on tumor proliferation and migration via the pyroptosis pathway in A549 and H1299 cells (36). The results of our study were aligned with these findings. The BTK gene (Bruton tyrosine kinase) was originally known as an essential target for B cell development and in the treatment of B cell malignancies (37). The function of BTK in LUAD remains unknown yet. Unlike BTK has a lower expression in LUAD than normal tissues (38). Recently, BTK has been considered an oncogenic target in solid tumors (39). Anti-BTK targeted drug, Ibrutinib, fails to achieve ideally protective efficacy in LUAD, whereas Auranofin enhances Ibrutinib’s activity in EGFR-mutant LUAD (40,41). Overexpression of p65-BTK groups, T790M-mutant, or erlotinib-resistant groups has been verified to selectively gain more benefits from BTK tyrosine kinase inhibitors (42,43). Meanwhile, a bioinformatical study reported that lower expression of BTK might be a protective predictor of LUAD (44). Controversially, BTK has been found as a tumor suppressor of p53-dependent senescence and apoptosis in other aspects (45,46). We found that BTK might play a role as a protective element related to pyroptosis in this prognostic study. The TUBB6 gene (tubulin beta 6 class V) encodes one type of β-tubulin, a structural subunit of microtubular α/β-heterodimers, which contains several individual isotypes with regulatory expression and distribution characteristics varying in mitotic and tumoral tissues (47,48). TUBB3, paralog of TUBB6, is the main constituent of tubulin and microtubule, inferior and superior structures of cytoskeleton respectively (49). Microtubule signifies division and growth in LUAD, and this is identified as the target of microtubule stabilizer, as known as taxanes (50). Tubulin-binding agents (TBAs) have shown early anti-tumor effects, mainly via apoptosis (51). Simultaneously, TBAs have been approved for their great clinical value as a single agent or regime combination with chemotherapy in advanced non-small cell lung cancer (NSCLC) (52,53). The advent of resistance to TBAs comes to a new challenge (54). The expression of TUBB3 has been reported as greatly increased in NSCLC and could be a prognostic and predictive factor of resistance to TBAs (55). Expression of TUBB6 is largely decreased in most tumors, including LUAD; however, its direct mechanism has yet to be clarified (56). Our results highlight that TUBB6 might provide another perspective towards prognostics in LUAD therapeutics. The MST1/MSP gene (macrophage stimulating 1/macrophage stimulating protein) is a secreted ligand mostly generated from the liver and activates its effect by binding to transmembrane tyrosine kinase receptor RON (recepteur d’origine nantais) mainly on the macrophage. The MSP-RON signaling pathway mediates inflammation and immune escape in physics (57). RON shares MET-like domain and function of ignition the tumorigenesis in LUAD (58). MSP-RON signaling pathway subsequently activates RAS-ERK and PI3K-Akt pathways to achieve development, migration, angiogenesis and chemoresistance in tumor cell (59). Whereas, the expression of RON in LUAD is not so highly distinct as breast, colon or pancreatic cancers, so another observation will be needed (60). Abrogation of RON expression resulted in a significant loss in viability and motility in NSCLC (61). Functional MSP inducing RON phosphorylation increases motility in H596, yet proliferation or apoptosis has not been observed (62). The MST1 gene has been identified as a novel prognostic mitogen in lung squamous cell carcinoma (63). In our study, MST1 was identified as a protective factor. Similarly, the same conclusions were drawn from the finding that the locating region of MST1 and RON coding genes sparks tumor suppressor activity and undergoes the frequent loss of heterozygosity (61).
Regulation of immune response was cardinally clustered in the pyroptosis-related 4-gene signature of LUAD. In addition, a previous study indicated an oncogenic role for GSDMD, one of the cardinal components of pyroptosis, in LUAD. On this basis, we considered poor outcome of LUAD might be attributed to coaction of the fractions of TME, and the immune landscape was subsequently analyzed through CIBERSORT. Among 7 high proportions of immunocytes between the landscapes, several cell lines have been reported as associated with malignant prospects in LUAD. A high concentration of plasma cell infiltration was confirmed as relating to the least favorable prognosis in LUAD (64). Besides, BTK is down-regulated as a transition from mature B cells to plasma cells (65). Abundant NK cell activated density is correlated with poor prognosis, and BTK is required for NK cell development (66,67). Macrophage presence is correlated with poor prognosis, since tumor-associated macrophages generally fuel pro-tumorigenic participation such as metastasis, angiogenesis, and immunosuppression (68,69). Additionally, high neutrophil proportion indicates a higher risk of LUAD (70).
The pyroptosis-related gene signature in this study holds some promising properties for in-depth research and long-term application in LUAD. The predictive pyroptosis-related gene signature would also be able to indicate prospective immune-related therapies in LUAD.
Conclusions
In conclusion, this study identified 26 DEGs with comprehensive bioinformatics analysis, provided their molecular mechanisms, and selected the potential biomarkers, MKI67, BTK, MST1, and TUBB6, which were grouped to predict progression of LUAD. However, further experiments in vitro and in vivo are required to validate the characteristics of these screened genes and pathways in pyroptosis progression in LUAD. In the future, we will continue to focus on pyroptosis function in LUAD through clinical and experimental studies.
Acknowledgments
We are grateful for the free access of TCGA and GEO databases.
Funding: This work was supported by the National Key R&D Program of China (2018YFC1705102).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-86/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-86/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Inage T, Nakajima T, Yoshino I, et al. Early Lung Cancer Detection. Clin Chest Med 2018;39:45-55. [Crossref] [PubMed]
- Kayagaki N, Dixit VM. Rescue from a fiery death: A therapeutic endeavor. Science 2019;366:688-9. [Crossref] [PubMed]
- Gao J, Qiu X, Xi G, et al. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non-small cell lung cancer. Oncol Rep 2018;40:1971-84. [Crossref] [PubMed]
- Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018;25:486-541. [Crossref] [PubMed]
- Jorgensen I, Rayamajhi M, Miao EA. Programmed cell death as a defence against infection. Nat Rev Immunol 2017;17:151-64. [Crossref] [PubMed]
- Fang Y, Tian S, Pan Y, et al. Pyroptosis: A new frontier in cancer. Biomed Pharmacother 2020;121:109595. [Crossref] [PubMed]
- Chen X, He WT, Hu L, et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res 2016;26:1007-20. [Crossref] [PubMed]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007;35:495-516. [Crossref] [PubMed]
- Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015;526:660-5. [Crossref] [PubMed]
- Kayagaki N, Stowe IB, Lee BL, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015;526:666-71. [Crossref] [PubMed]
- Teng JF, Mei QB, Zhou XG, et al. Polyphyllin VI Induces Caspase-1-Mediated Pyroptosis via the Induction of ROS/NF-κB/NLRP3/GSDMD Signal Axis in Non-Small Cell Lung Cancer. Cancers (Basel) 2020;12:193. [Crossref]
- Wang Y, Gao W, Shi X, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017;547:99-103. [Crossref] [PubMed]
- Zhang CC, Li CG, Wang YF, et al. Chemotherapeutic paclitaxel and cisplatin differentially induce pyroptosis in A549 lung cancer cells via caspase-3/GSDME activation. Apoptosis 2019;24:312-25. [Crossref] [PubMed]
- Lin W, Chen Y, Wu B, et al. Identification of the pyroptosis-related prognostic gene signature and the associated regulation axis in lung adenocarcinoma. Cell Death Discov 2021;7:161. [Crossref] [PubMed]
- Liu LP, Lu L, Zhao QQ, et al. Identification and Validation of the Pyroptosis-Related Molecular Subtypes of Lung Adenocarcinoma by Bioinformatics and Machine Learning. Front Cell Dev Biol 2021;9:756340. [Crossref] [PubMed]
- Zhang G, Yan Z. A New Definition of Pyroptosis-Related Gene Markers to Predict the Prognosis of Lung Adenocarcinoma. Biomed Res Int 2021;2021:8175003. [Crossref] [PubMed]
- Li L, Jiang M, Qi L, et al. Pyroptosis, a new bridge to tumor immunity. Cancer Sci 2021;112:3979-94. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol 2013;31:3987-96. [Crossref] [PubMed]
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. [Crossref] [PubMed]
- Calvayrac O, Pradines A, Pons E, et al. Molecular biomarkers for lung adenocarcinoma. Eur Respir J 2017;49:1601734. [Crossref] [PubMed]
- Tang R, Xu J, Zhang B, et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol 2020;13:110. [Crossref] [PubMed]
- Chen L, Weng B, Li H, et al. A thiopyran derivative with low murine toxicity with therapeutic potential on lung cancer acting through a NF-κB mediated apoptosis-to-pyroptosis switch. Apoptosis 2019;24:74-82. [Crossref] [PubMed]
- Wang WJ, Chen D, Jiang MZ, et al. Downregulation of gasdermin D promotes gastric cancer proliferation by regulating cell cycle-related proteins. J Dig Dis 2018;19:74-83. [Crossref] [PubMed]
- Li Q, Chen L, Dong Z, et al. Piperlongumine analogue L50377 induces pyroptosis via ROS mediated NF-κB suppression in non-small-cell lung cancer. Chem Biol Interact 2019;313:108820. [Crossref] [PubMed]
- Tapeinos C, Pandit A. Physical, Chemical, and Biological Structures based on ROS-Sensitive Moieties that are Able to Respond to Oxidative Microenvironments. Adv Mater 2016;28:5553-85. [Crossref] [PubMed]
- Le X, Mu J, Peng W, et al. DNA methylation downregulated ZDHHC1 suppresses tumor growth by altering cellular metabolism and inducing oxidative/ER stress-mediated apoptosis and pyroptosis. Theranostics 2020;10:9495-511. [Crossref] [PubMed]
- Wu M, Wang Y, Yang D, et al. A PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma. EBioMedicine 2019;41:244-55. [Crossref] [PubMed]
- Cuylen S, Blaukopf C, Politi AZ, et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 2016;535:308-12. [Crossref] [PubMed]
- Menon SS, Guruvayoorappan C, Sakthivel KM, et al. Ki-67 protein as a tumour proliferation marker. Clin Chim Acta 2019;491:39-45. [Crossref] [PubMed]
- Chirieac LR. Ki-67 expression in pulmonary tumors. Transl Lung Cancer Res 2016;5:547-51. [Crossref] [PubMed]
- Li Z, Li F, Pan C, et al. Tumor cell proliferation (Ki-67) expression and its prognostic significance in histological subtypes of lung adenocarcinoma. Lung Cancer 2021;154:69-75. [Crossref] [PubMed]
- Wang F, Liu W, Ning J, et al. Simvastatin Suppresses Proliferation and Migration in Non-small Cell Lung Cancer via Pyroptosis. Int J Biol Sci 2018;14:406-17. [Crossref] [PubMed]
- Burger JA, Wiestner A. Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat Rev Cancer 2018;18:148-67. [Crossref] [PubMed]
- Yang X, Zheng Y, Han Z, et al. Functions and clinical significance of KLRG1 in the development of lung adenocarcinoma and immunotherapy. BMC Cancer 2021;21:752. [Crossref] [PubMed]
- Molina-Cerrillo J, Alonso-Gordoa T, Gajate P, et al. Bruton's tyrosine kinase (BTK) as a promising target in solid tumors. Cancer Treat Rev 2017;58:41-50. [Crossref] [PubMed]
- Hong D, Rasco D, Veeder M, et al. A Phase 1b/2 Study of the Bruton Tyrosine Kinase Inhibitor Ibrutinib and the PD-L1 Inhibitor Durvalumab in Patients with Pretreated Solid Tumors. Oncology 2019;97:102-11. [Crossref] [PubMed]
- Hu J, Zhang H, Cao M, et al. Auranofin Enhances Ibrutinib's Anticancer Activity in EGFR-Mutant Lung Adenocarcinoma. Mol Cancer Ther 2018;17:2156-63. [Crossref] [PubMed]
- Gao W, Wang M, Wang L, et al. Selective antitumor activity of ibrutinib in EGFR-mutant non-small cell lung cancer cells. J Natl Cancer Inst 2014;106:dju204. [Crossref] [PubMed]
- Giordano F, Vaira V, Cortinovis D, et al. p65BTK is a novel potential actionable target in KRAS-mutated/EGFR-wild type lung adenocarcinoma. J Exp Clin Cancer Res 2019;38:260. [Crossref] [PubMed]
- Bi KW, Wei XG, Qin XX, et al. BTK Has Potential to Be a Prognostic Factor for Lung Adenocarcinoma and an Indicator for Tumor Microenvironment Remodeling: A Study Based on TCGA Data Mining. Front Oncol 2020;10:424. [Crossref] [PubMed]
- Rada M, Barlev N, Macip S. BTK: a two-faced effector in cancer and tumour suppression. Cell Death Dis 2018;9:1064. [Crossref] [PubMed]
- Althubiti M, Rada M, Samuel J, et al. BTK Modulates p53 Activity to Enhance Apoptotic and Senescent Responses. Cancer Res 2016;76:5405-14. [Crossref] [PubMed]
- Janke C, Magiera MM. The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol 2020;21:307-26. [Crossref] [PubMed]
- Lopes D, Maiato H. The Tubulin Code in Mitosis and Cancer. Cells 2020;9:2356. [Crossref] [PubMed]
- Shao Q, Yang T, Huang H, et al. Uncoupling of UNC5C with Polymerized TUBB3 in Microtubules Mediates Netrin-1 Repulsion. J Neurosci 2017;37:5620-33. [Crossref] [PubMed]
- Gonçalves A, Braguer D, Kamath K, et al. Resistance to Taxol in lung cancer cells associated with increased microtubule dynamics. Proc Natl Acad Sci U S A 2001;98:11737-42. [Crossref] [PubMed]
- Haider K, Rahaman S, Yar MS, et al. Tubulin inhibitors as novel anticancer agents: an overview on patents (2013-2018). Expert Opin Ther Pat 2019;29:623-41. [Crossref] [PubMed]
- Borisy G, Heald R, Howard J, et al. Microtubules: 50 years on from the discovery of tubulin. Nat Rev Mol Cell Biol 2016;17:322-8. [Crossref] [PubMed]
- Hardin C, Shum E, Singh AP, et al. Emerging treatment using tubulin inhibitors in advanced non-small cell lung cancer. Expert Opin Pharmacother 2017;18:701-16. [Crossref] [PubMed]
- Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer 2010;10:194-204. [Crossref] [PubMed]
- Sève P, Dumontet C. Is class III beta-tubulin a predictive factor in patients receiving tubulin-binding agents? Lancet Oncol 2008;9:168-75. [Crossref] [PubMed]
- Cucchiarelli V, Hiser L, Smith H, et al. Beta-tubulin isotype classes II and V expression patterns in nonsmall cell lung carcinomas. Cell Motil Cytoskeleton 2008;65:675-85. [Crossref] [PubMed]
- Huang L, Fang X, Shi D, et al. MSP-RON Pathway: Potential Regulator of Inflammation and Innate Immunity. Front Immunol 2020;11:569082. [Crossref] [PubMed]
- Angeloni D, Danilkovitch-Miagkova A, Ivanov SV, et al. Gene structure of the human receptor tyrosine kinase RON and mutation analysis in lung cancer samples. Genes Chromosomes Cancer 2000;29:147-56. [Crossref] [PubMed]
- Yao HP, Zhou YQ, Zhang R, et al. MSP-RON signalling in cancer: pathogenesis and therapeutic potential. Nat Rev Cancer 2013;13:466-81. [Crossref] [PubMed]
- Wagh PK, Peace BE, Waltz SE. Met-related receptor tyrosine kinase Ron in tumor growth and metastasis. Adv Cancer Res 2008;100:1-33. [Crossref] [PubMed]
- Kanteti R, Krishnaswamy S, Catenacci D, et al. Differential expression of RON in small and non-small cell lung cancers. Genes Chromosomes Cancer 2012;51:841-51. [Crossref] [PubMed]
- Willett CG, Wang MH, Emanuel RL, et al. Macrophage-stimulating protein and its receptor in non-small-cell lung tumors: induction of receptor tyrosine phosphorylation and cell migration. Am J Respir Cell Mol Biol 1998;18:489-96. [Crossref] [PubMed]
- Van de Laar E, Clifford M, Hasenoeder S, et al. Cell surface marker profiling of human tracheal basal cells reveals distinct subpopulations, identifies MST1/MSP as a mitogenic signal, and identifies new biomarkers for lung squamous cell carcinomas. Respir Res 2014;15:160. [Crossref] [PubMed]
- Kurebayashi Y, Emoto K, Hayashi Y, et al. Comprehensive Immune Profiling of Lung Adenocarcinomas Reveals Four Immunosubtypes with Plasma Cell Subtype a Negative Indicator. Cancer Immunol Res 2016;4:234-47. [Crossref] [PubMed]
- Jumaa H, Hendriks RW, Reth M. B cell signaling and tumorigenesis. Annu Rev Immunol 2005;23:415-45. [Crossref] [PubMed]
- Chen Y, Chen H, Mao B, et al. Transcriptional Characterization Of The Tumor Immune Microenvironment And Its Prognostic Value For Locally Advanced Lung Adenocarcinoma In A Chinese Population. Cancer Manag Res 2019;11:9165-73. [Crossref] [PubMed]
- Weber ANR, Bittner Z, Liu X, et al. Bruton's Tyrosine Kinase: An Emerging Key Player in Innate Immunity. Front Immunol 2017;8:1454. [Crossref] [PubMed]
- Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014;41:49-61. [Crossref] [PubMed]
- Cheng N, Bai X, Shu Y, et al. Targeting tumor-associated macrophages as an antitumor strategy. Biochem Pharmacol 2021;183:114354. [Crossref] [PubMed]
- Sun L, Jiang G, Gonzalez-Rivas D, et al. An individualized immune prognostic signature in lung adenocarcinoma. Cancer Cell Int 2020;20:156. [Crossref] [PubMed]
(English Language Editor: J. Jones)


