
Comprehensive analysis of aneuploidy status and its effect on the efficacy of EGFR-TKIs in lung cancer
Introduction
Lung cancer, the leading cause of cancer death globally, arises due to the acquisition of somatic alterations in patients’ genomes, which alter the function of key cancer genes. A number of these alterations are implicated as determinants of treatment response in clinical practice (1). Presently, lung cancer driver mutations and mutational signatures have been described in detail (2,3), and numerous clinical trials have demonstrated that targeted therapies can delay the progression of the disease in patients. Targeted drugs have achieved considerable clinical success and significantly extended the progression-free survival (PFS) and overall survival (OS) in lung cancer patients with driver gene mutations. However, accurate and effective evaluation of the patient response to targeted drugs has always been a difficult problem for clinicians due to tumor heterogeneity and individual differences among patients.
Biomarkers are crucial when selecting the best therapy; that is, an identifiable, tumor-specific feature that can predict drug response. Aneuploidy-driven phenotypes are caused by gene copy number changes. The classical definition of aneuploidy is the numerical aberrations of whole chromosomes. Recent studies have suggested that aneuploidy is a context-dependent, cancer-type-specific, oncogenic event that may have clinical relevance as a prognostic marker and potential therapeutic target (4,5). Aneuploidy is a hallmark of most cancers and is associated with increased malignancy, tumor recurrence, and drug resistance (6-8). Indeed, roughly two out of three human tumors display aneuploidy, and the incidence of DNA aneuploidy in small cell lung cancer is 77.8%, which is higher than any other histological type of lung cancer (9-11). Additionally, increasing aneuploidy levels are generally correlated with later tumor stages (12). Emerging evidence indicates that aneuploidy is a novel driving force in tumorigenesis and is associated with prognosis beyond strong predictors (13). The prognostic value of aneuploidy has long been demonstrated for several indications (14,15), with high levels of aneuploidy being associated with poorer prognosis in the vast majority of cases. Previous study has confirmed that aneuploidy qualifies as a predictive biomarker for benefit from adjuvant docetaxel (16). Tumor aneuploidy may also be a useful biomarker for predicting which patients are most likely to benefit from immunotherapy (17). Epidermal growth factor receptor (EGFR) is a tumor driver gene of lung cancer, which guides tumorigenesis and growth by activating the EGFR signaling pathway (18). However, an important question is whether aneuploidy can also inform treatment decisions for targeted therapy. The mechanism through which aneuploidy influences cancer progression and whether degree of aneuploidy can be implemented clinically to inform the care of patients with cancer remain unclear.
In our study, aneuploidy status and EGFR mutation abundance were used to evaluate the clinical efficacy of patients with lung cancer. The results support a strong correlation between aneuploidy status and patient efficacy, with euploid patients having better clinical efficacy. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-73/rc).
Methods
Patient specimen acquisition
We retrospectively investigated the medical records of lung cancer patients who underwent EGFR mutation and mutation abundance examinations in tumor tissues by next-generation sequencing (NGS) of an eight-gene panel and fluorescence in situ hybridization (FISH) from July 2018 to June 2020. The clinical data of all patients were obtained from the electronic medical record database of Henan Cancer Hospital. The present study included 63 lung cancer patients who received NGS assessment and FISH for molecularly matched therapy and follow-up. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocols were approved by the ethics committee of Henan Cancer Hospital (No. 2021-KY-0184-002). Written informed consent was obtained from all individuals included in the study.
DNA extraction and qualification, and target gene sequencing
Genomic DNA was extracted from tumor formalin-fixed paraffin-embedded (FFPE) tissues using a DNeasy Blood & Tissue Kit (Qiagen Inc., USA). The KAPA Hyper Prep Kit (Kapa Biosystems, USA) was utilized for DNA library preparation as a versatile reagent kit adapted to the Illumina platform. For hybridization enrichment, customized xGen lockdown probes (Integrated DNA Technologies, USA) were applied. The probes panel was designed to target eight tumor-specific genes. All procedures were conducted according to the manufacturers’ protocols.
FISH experiments and the detection result of the determination criteria
All FISH tests were conducted in the Department of Pathology at the Henan Cancer Center. FISH was carried out on FFPE sections using the ALK (anaplastic lymphoma kinase) Break Apart Detection kit (Guangzhou LBP Medicine Science & Technology Co., Ltd., China) according to manufacturer’s instructions. The GSP ALK 5’ probe (Spectrum Green) and the GSP LAF4 3’ probe (Spectrum RED) were labeled, hybridized, and evaluated along with the standard controls. FISH signals were evaluated independently by two technicians who were blinded to the patient’s history and histologic findings. All samples were examined by pathologists to identify the tumor cell-enriched areas, which were marked on the underside of the slides with a diamond-tipped scribe. The percentage of tumor cells in each case was over 60%. The presence of separated green-red signals (>2 signal diameters) or individual red signals in tumor cells was considered FISH-positive, while FISH-negative was defined as overlapping red-green signals (slightly yellow).
The criteria for detecting ALK status in the sample were that at least 50 tumor cells were observed. If >25 cells out of 50 (>25/50 or >50%) were positive, the sample was considered positive. If 5–25 cells out of 50 (10–50%) were positive, the sample was considered equivocal, and the slide was evaluated by the second reader who selected 50 additional nuclei. In this case, the sample was considered positive if >15 of the accumulated 100 cells had a separation signal (≥15%).
Study endpoints
The primary endpoint of this study was to determine the association between PFS and mutation abundance of eligible patients after EGFR- tyrosine kinase inhibitor (TKI) therapy. PFS was defined as the time from initiating the administration of oral EGFR-TKI to disease progression. The secondary endpoints included the objective response rate (ORR), as well as the association between PFS and age, sex, and whether EGFR-TKI was administered as the first-line treatment. The therapeutic effect of EGFR-TKI was evaluated according to the Response Evaluation Criteria In Solid Tumors: complete response (CR), which signified the disappearance of all target lesions; partial response (PR), which denoted at least a 30% decrease in the sum of the longest diameter of target lesions with the baseline sum of the longest diameter as the reference; and progressive disease (PD), defined as the ratio between the longest diameter sum of the target lesion and the longest diameter of the smallest target lesion recorded after beginning the administration of oral EGFR-TKI, which increased by at least 20% or the presence of one or more new lesions. The ORR was defined as achieving CR or PR.
Statistical methods
All statistical analyzes were performed using GraphPad Prism version 7.0 (GraphPad Software, Inc., USA). Kaplan-Meier and Cox survival regression models were used to analyze the influence of age, gender, aneuploidy status, and EGFR mutation abundance on survival and prognosis. P<0.05 was regarded as statistically significant.
Results
Patient characteristics
A total of 291 patients diagnosed with lung cancer were analyzed by both NGS and FISH methods. Among them, 63 (21.6%) patients were treated according to the molecular testing and followed-up constantly. The clinical and pathological features of the lung cancer patients are summarized in Table 1. There were 23 males (36.5%) and 40 females (63.5%), 20.6% (13/63) had a smoking history, and 22.2% (14/63) were ≥65 years old. The median age of the patients was 57 years (ranging from 31 to 76 years). There were 60 patients with adenocarcinoma (95.2%), one with squamous cell carcinoma (1.6%), and two with non-small cell lung cancer (NSCLC) (3.2%).
Table 1
| Characteristics | All patients (n=63) |
|---|---|
| Age | |
| Median [range] | 57 [31–76] |
| <65 | 49 (77.8) |
| ≥65 | 14 (22.2) |
| Sex, n (%) | |
| Male | 23 (36.5) |
| Female | 40 (63.5) |
| Smoking history, n (%) | |
| Ever smoker | 13 (20.6) |
| Never smoker | 50 (79.4) |
| Pathology type, n (%) | |
| Adenocarcinoma | 60 (95.2) |
| Squamous | 1 (1.6) |
| NSCLC | 2 (3.2) |
| Stage, n (%) | |
| I–III | 15 (23.8) |
| IV | 48 (76.2) |
NSCLC, non-small cell lung cancer.
PFS was correlated with EGFR mutation abundance after EGFR-TKI therapy
The median PFS was 10.0 months (range, 1.0–33.0 months), with an overall disease control rate of 42.4%. In order to establish the accurate cutoff value for distinguishing EGFR mutation abundance and thus provide useful evidence for clinical practice, combining the median PFS data of patients who received EGFR-TKI orally were analyzed by stratification.
Stratified analyses were conducted by combining the median PFS data of patients given EGFR-TKI orally. We respectively tried to use 10%, 20%, 30%, 40%, or 50% as the cutoff values to analyze the medium PFS of the high- and low-abundance groups. The results revealed that when 10%, 20%, or 30% were used as the cutoff values, the medium PFS achieved a statistically significant therapeutic effect in >10%, 20%, and 30% compared to <10%, 20%, and 30% mutation abundances, respectively (Table S1). The medium PFS was most significant (11.0 vs. 8.0 months) when an EGFR mutation abundance value of 25% was used as the cutoff value. Although the median PFS increased initially with an increase in the abundance of the EGFR mutation, the median PFS did not continue to rise when the mutation abundance grew more than 30%.
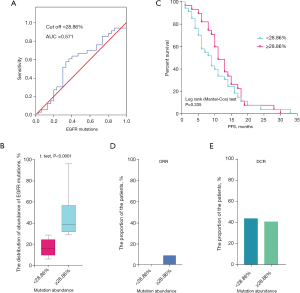
Furthermore, we estimated the accurate cut-off value of EGFR mutation abundance by ROC (receiver operating characteristic) analysis in the 63 patients. We found that the cut-off value was 28.86% and the area under the curve (AUC) was 0.571 (0.426–0.716) for EGFR mutation abundance (Figure 1A). When the cut-off value of the abundance of EGFR mutations was ≥28.86%, the median mutation abundance was about 40% (Figure 1B). Also, Kaplan-Meier analysis showed that a cut-off value of 28.86% did not exhibit a significant difference between the PFS duration and EGFR mutation abundance (Figure 1C). Simultaneously, we investigated the difference in clinical efficacy between patients with an EGFR mutation abundance <28.86% and ≥28.86%, and found that patients with an EGFR mutation abundance ≥28.86% had slightly a higher ORR and similar DCR (Figure 1D,1E).
PFS was correlated with aneuploidy status after EGFR-TKI therapy
At the same time, all 63 cases also received FISH assessment for aneuploidy status. The median progression-free survival (mPFS) of euploid patients was 9.0 months (40 cases), and that of patients with aneuploidy was 10.0 months (23 cases) (Figure 2A). To further investigate whether euploidy and aneuploidy affected the patients’ PFS, Kaplan-Meier analysis was employed and showed no significant difference between euploid and aneuploid patients (Figure 2B).
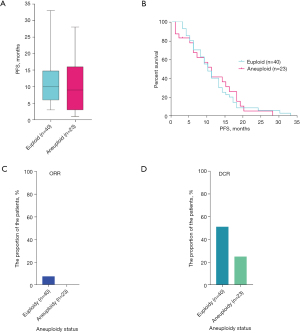
Furthermore, we explored the dissimilitude in clinical efficacy between euploid and aneuploid patients, and found that euploid patients displayed a higher ORR and DCR. The ORRs of euploid and aneuploid patients were 7.7% and 0%, and the DCRs were 51.3% and 25.0%, respectively (Figure 2C,2D). The results showed that the PFS of patients diagnosed as euploidy was actually higher than that of patients diagnosed with aneuploidy, and was related to both ORR and DCR, suggesting that patients diagnosed with euploidy had a better clinical efficacy.
Association between aneuploidy status, EGFR mutation abundance, and clinical efficacy
We also further elucidated whether tumor aneuploidy status and the abundance of EGFR mutations together affect PFS, and found that there were significant differences between both aneuploidy status and EGFR mutation abundance in the 63 patients. The median EGFR mutation abundance in patients with euploidy was 25.51%, while that in aneuploid patients was 28.59% (Figure 3A). Two-way analysis of variance was also used to assess the effects of EGFR mutation abundance and tumor aneuploidy status on the patients’ PFS. In patients with EGFR mutation abundance <28.86%, the euploid patients was slightly more significant than the aneuploid patients. However, in patients with EGFR mutation abundance >28.86%, there was no significant difference between euploid and aneuploid patients (Figure 3B). When the EGFR mutation abundance was <28.86%, the mPFS of patients with euploidy and aneuploidy were 9 and 11 months, respectively. Meanwhile, for patients with an EGFR mutation abundance >28.86%, the mPFS were 11 and 10 months, respectively.
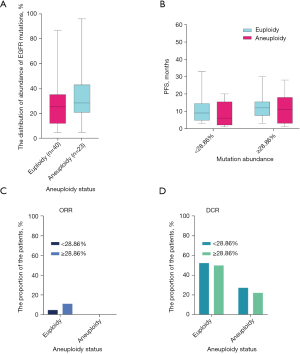
We also assessed the dissimilitude of clinical efficacy between both EGFR mutation abundance and aneuploidy status. Euploid patients exhibited higher ORR and DCR than aneuploid patients, regardless of EGFR mutation abundance (Figure 3C,3D). Next, we also conducted univariate analysis for the general characteristics of age, gender, and smoking history through Kaplan-Meier survival analyses. The results showed that these factors were not associated with the median PFS after EGFR-TKI (Table S2).
Discussion
Cancer is driven by multiple types of genetic alterations, ranging in size from point mutations to whole-chromosome gains and losses, known as aneuploidy. Chromosome instability, the process that gives rise to aneuploidy and can promote tumorigenesis by increasing genetic heterogeneity and promoting tumor evolution. In the recent cancer genome literature, this definition has been extended to include gains or losses of chromosome arms (19,20). Unlike ‘aneuploidy’, the term ‘focal copy number alterations (focal CNAs)’ is usually used to describe smaller copy number changes that encompass fewer genes. Although this qualitative definition of aneuploidy is operationally convenient, it is ambiguous (4). The consequence of aneuploidy for the genome is mostly limited to alterations in gene copy numbers. It is generally believed that changes in the copy number of specific genes are responsible for the increased fitness of cells harboring specific aneuploidies (21-26).
Recent studies have further suggested that aneuploidies are largely driven by the cumulative effects of oncogenes and tumor suppressor genes that reside within the aberrant chromosome (27,28). A key feature of aneuploid cells is that they often provoke genomic instability (29-31). A single episode of genomic instability generates multiple subclones that attain clonal dominance at different rates and under various selective pressures (32), and clonal suppression and recrudescence appear to correspond to drug sensitivity and resistance. Since therapeutic agents can be regarded as new selection pressures, aneuploidy might differentially influence the therapeutic response and tumor relapse in ways similar to their impact on tumor evolution (33). Aneuploidy can have profound impact on therapy by accelerating tumorigenesis and the outgrowth of resistant clones followed by tumor release. Targeting the aneuploid state, specific aneuploidy drivers or specific aneuploidy passengers have been shown to be useful in selectively killing aneuploid cells. Therapeutically exploiting aneuploidy will likely depend on the mechanism and level of aneuploidy tolerance in aneuploid cells. Since aneuploidy tolerance might be a bottleneck for increasing karyotypic divergence, low divergence might predict higher sensitivity toward such therapeutic strategies.
Aneuploidy is associated with tumor progression and poor prognosis (16). Aneuploidy and SCNA (somatic copy number alteration) levels in cancers have been shown to positively correlate with mutation load and cell proliferation (11,25). By determining the copy number of specific chromosomes in single cells, it was shown that the chromosomal content within certain cell populations varies over time. Aneuploidy is usually quantified by measuring intracellular DNA content or chromosome structure and number (16). Therefore, aneuploidy can be considered as a biomarker for evaluating clinical efficacy.
In contrast, single-cell methods, such as FISH, provide a more accurate measure of focal SCNAs. In order to examine the correlation between focal SCNAs and clinical outcomes, we simultaneously used NGS and FISH to obtain the clinical information of patients. Our results demonstrated that patients diagnosed with euploidy had better clinical efficacy, as well as a higher ORR and DCR (Figure 2C,2D). Two-way analysis of variance also showed that the mPFS of euploid patients was higher than that of aneuploid patients, and the ORR and DCR of euploid patients were better than those of aneuploid patients (Figure 3C,3D). Our results support the strong correlation between clinical efficacy and cancer aneuploidy status.
The success of precision medicine depends on our ability to effectively translate genomic data into actionable, customized prognosis and treatment regimens for individual patients. Valid predictive factors of the efficacy of EGFR-TKIs are important for selecting patients who may benefit more from EGFR-TKI treatment. Several previous studies have shown that EGFR amplification and overexpression are often associated with aneuploidy and tumor cell proliferation, and that the relative abundance of EGFR mutations can predict the degree of benefit from EGFR-TKI therapy (34,35). However, our study confirmed that the PFS of patients was not closely related to the mutation abundance of the EGFR gene after treatment with EGFR-TKI (Figure 3B,3D). In comparison to aneuploidy status, treatment with EGFR-TKI in patients with high EGFR mutation abundance did not achieve more benefits in terms of ORR and DCR, while 28.86% was the best cutoff value to separate between low and high EGFR mutant abundance (Figure 1D,1E). Our results support that clinical efficacy has no correlation with EGFR mutation abundance.
The clinical development of molecularly targeted cancer therapies remains a huge challenge. The FISH technique plays a leading role in diagnostic pathology for its single-cell analysis, and has provided crucial information regarding genomic variations in malignant cells. Although aneuploidy status can relatively quickly be determined by FISH, this technique has several limitations. The main drawback of is the fact that most tumors cannot be scored in an automated fashion, which renders FISH extremely labor intensive, limiting its potential use in a clinical setting. Secondly, FISH can be performed on interphase nuclei, but can only analyze a limited number of loci. It seems probable that such single-cell, NGS approaches to define tumor aneuploidy status will become more prevalent as costs decrease and technology improves. In recent years, aneuploidy can readily be detected using multiple technologies, including various conventional and molecular cytogenetic methods, FISH (36), single-nucleotide polymorphism arrays (SNP array), as well as genome-wide DNA and RNA (ribonucleic acid) sequencing (4). Furthermore, other molecular cytogenetic methodologies, such as chromosome specific FISH karyotyping and comparative genomic hybridization, have also helped in the detection of cryptic genetic changes in cancer.
In our study, aneuploidy status and EGFR mutation abundance were used to evaluate clinical efficacy of patients with lung cancer. The results support a strong correlation between aneuploidy status and clinical efficacy, and euploid patients exhibited a higher ORR and DCR.
Conclusions
In summary, our data suggests that assays of tumor aneuploidy status might be useful for determining which patients are most likely to respond to therapies based on EGFR-TKIs. Information on aneuploidy status can be derived from NGS analysis and FISH performed on patient tumors. Our results suggest that important clues to the progression of lung cancer lie in the aneuploidy status.
Acknowledgments
The authors would like to thank Shanghai Tongshu Biotechnology Co., Ltd. for their technical support. This manuscript was submitted as a pre-print at https://www.researchsquare.com/article/rs-845098/v1
Funding: This project was funded by the big data construction of precision medicine for common malignant tumors such as lung cancer, a major science and technology project of Henan Province (project No. 161100311500).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-73/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-73/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-73/coif). All authors report that this work received technical support from Shanghai Tongshu Biotechnology Co. Ltd. The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016;166:740-54. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Martínez-Jiménez F, Muiños F, Sentís I, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20:555-72. [Crossref] [PubMed]
- Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet 2020;21:44-62. [Crossref] [PubMed]
- Kristensen GB, Kildal W, Abeler VM, et al. Large-scale genomic instability predicts long-term outcome for women with invasive stage I ovarian cancer. Ann Oncol 2003;14:1494-500. [Crossref] [PubMed]
- Carter SL, Eklund AC, Kohane IS, et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 2006;38:1043-8. [Crossref] [PubMed]
- McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle 2009;8:3262-6. [Crossref] [PubMed]
- Sotillo R, Schvartzman JM, Socci ND, et al. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 2010;464:436-40. [Crossref] [PubMed]
- Duijf PH, Schultz N, Benezra R. Cancer cells preferentially lose small chromosomes. Int J Cancer 2013;132:2316-26. [Crossref] [PubMed]
- Ariyoshi I. Characteristics and problems of flow cytometric nuclear DNA content analysis in lung cancer using bronchoscopically-obtained specimens. Nihon Kyobu Shikkan Gakkai Zasshi 1992;30:1704-10. [PubMed]
- Nakanishi K, Hiroi S, Kawai T, et al. Argyrophilic nucleolar-organizer region counts and DNA status in bronchioloalveolar epithelial hyperplasia and adenocarcinoma of the lung. Hum Pathol 1998;29:235-9. [Crossref] [PubMed]
- Laubert T, Freitag-Wolf S, Linnebacher M, et al. Stage-specific frequency and prognostic significance of aneuploidy in patients with sporadic colorectal cancer--a meta-analysis and current overview. Int J Colorectal Dis 2015;30:1015-28. [Crossref] [PubMed]
- Li X, Cai W, Yang G, et al. Comprehensive Analysis of EGFR-Mutant Abundance and Its Effect on Efficacy of EGFR TKIs in Advanced NSCLC with EGFR Mutations. J Thorac Oncol 2017;12:1388-97. [Crossref] [PubMed]
- Tanaka A, Sueoka-Aragane N, Nakamura T, et al. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer 2012;75:89-94. [Crossref] [PubMed]
- Steinbeck RG, Heselmeyer KM, Auer GU. DNA ploidy in human colorectal adenomas. Anal Quant Cytol Histol 1994;16:196-202. [PubMed]
- Stopsack KH, Whittaker CA, Gerke TA, et al. Aneuploidy drives lethal progression in prostate cancer. Proc Natl Acad Sci U S A 2019;116:11390-5. [Crossref] [PubMed]
- Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355:eaaf8399. [Crossref] [PubMed]
- Ito K, Morise M, Wakuda K, et al. A multicenter cohort study of osimertinib compared with afatinib as first-line treatment for EGFR-mutated non-small-cell lung cancer from practical dataset: CJLSG1903. ESMO Open 2021;6:100115. [Crossref] [PubMed]
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676-689.e3. [Crossref] [PubMed]
- Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134-40. [Crossref] [PubMed]
- Ben-David U, Arad G, Weissbein U, et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun 2014;5:4825. [Crossref] [PubMed]
- Rutledge SD, Douglas TA, Nicholson JM, et al. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep 2016;6:22828. [Crossref] [PubMed]
- Pavelka N, Rancati G, Zhu J, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 2010;468:321-5. [Crossref] [PubMed]
- Kawakami M, Liu X, Dmitrovsky E. New Cell Cycle Inhibitors Target Aneuploidy in Cancer Therapy. Annu Rev Pharmacol Toxicol 2019;59:361-77. [Crossref] [PubMed]
- McGranahan N, Burrell RA, Endesfelder D, et al. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep 2012;13:528-38. [Crossref] [PubMed]
- Lee AJ, Endesfelder D, Rowan AJ, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 2011;71:1858-70. [Crossref] [PubMed]
- Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013;155:948-62. [Crossref] [PubMed]
- Sack LM, Davoli T, Li MZ, et al. Profound Tissue Specificity in Proliferation Control Underlies Cancer Drivers and Aneuploidy Patterns. Cell 2018;173:499-514.e23. [Crossref] [PubMed]
- Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013;494:492-6. [Crossref] [PubMed]
- Lamm N, Ben-David U, Golan-Lev T, et al. Genomic Instability in Human Pluripotent Stem Cells Arises from Replicative Stress and Chromosome Condensation Defects. Cell Stem Cell 2016;18:253-61. [Crossref] [PubMed]
- Ly P, Brunner SF, Shoshani O, et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat Genet 2019;51:705-15. [Crossref] [PubMed]
- Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood 2012;120:1067-76. [Crossref] [PubMed]
- Theillet C. Towards an inventory of oncogenic mutations in cancer. Bull Cancer 2010;97:1223-9. [Crossref] [PubMed]
- Konovalov NA, Asyutin DS, Shayhaev EG, et al. Molecular Biomarkers of Brain and Spinal Cord Astrocytomas. Acta Naturae 2019;11:17-27. [Crossref] [PubMed]
- Wang H, Zhang M, Tang W, et al. Mutation abundance affects the therapeutic efficacy of EGFR-TKI in patients with advanced lung adenocarcinoma: A retrospective analysis. Cancer Biol Ther 2018;19:687-94. [Crossref] [PubMed]
- Bakker B, van den Bos H, Lansdorp PM, et al. How to count chromosomes in a cell: An overview of current and novel technologies. Bioessays 2015;37:570-7. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


